8RPO
 
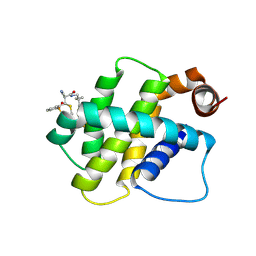 | | BFL1 in complex with a reversible covalent ligand | | Descriptor: | (~{E})-2-cyano-3-(2-dimethylphosphorylphenyl)-~{N}-[[1-[4-(trifluoromethyl)phenyl]cyclopropyl]methyl]prop-2-enamide, Bcl-2-related protein A1 | | Authors: | Hargreaves, D. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Identification and Evaluation of Reversible Covalent Binders to Cys55 of Bfl-1 from a DNA-Encoded Chemical Library Screen.
Acs Med.Chem.Lett., 15, 2024
|
|
6QVS
 
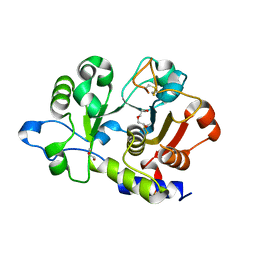 | | Unliganded structure of the human wild type Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6Gal1) | | Descriptor: | Beta-galactoside alpha-2,6-sialyltransferase 1, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Harrus, D, Glumoff, T. | | Deposit date: | 2019-03-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unliganded and CMP-Neu5Ac bound structures of human alpha-2,6-sialyltransferase ST6Gal I at high resolution.
J.Struct.Biol., 212, 2020
|
|
2YJ8
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-(4-iodophenyl)cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
2BPM
 
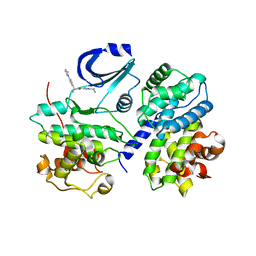 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-630529 | | Descriptor: | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL-3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN-1-YL)PHENYL]PROPANAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Brasca, M.G, Orsini, P, Traquandi, G, Longo, A, Nesi, M, Orzi, F, Piutti, C, Sansonna, P, Varasi, M, Vulpetti, A, Roletto, F, Alzani, R, Ciomei, M, Albanese, C, Pastori, W, Marsiglio, A, Pesenti, E, Fiorentini, F, Bischoff, J.R, Mercurio, C. | | Deposit date: | 2005-04-21 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2-Cyclin a as Antitumor Agents. 2. Lead Optimization
J.Med.Chem., 48, 2005
|
|
4KV0
 
 | |
6KR6
 
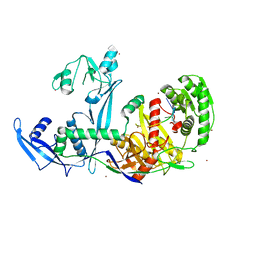 | | Crystal structure of Drosophila Piwi | | Descriptor: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | Authors: | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
2YJB
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-[4-(trifluoromethyl)phenyl]cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
5KMW
 
 | | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | | Descriptor: | Beta-lactamase Toho-1, OPEN FORM - PENICILLIN G, PENICILLIN G, ... | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Ginell, S.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex
to be published
|
|
2C69
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2C5V
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)-N-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE, ALA-ALA-ABA-ARG-SER-LEU-ILE-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-02 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2CCI
 
 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
4L1X
 
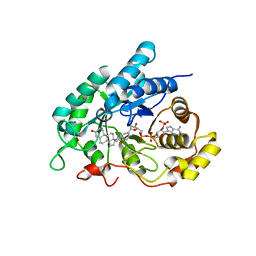 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 V54L Mutant in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
2CJM
 
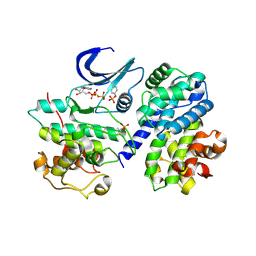 | | Mechanism of CDK inhibition by active site phosphorylation: CDK2 Y15p T160p in complex with cyclin A structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Welburn, J.P.I, Tucker, J, Johnson, T, Lindert, L, Morgan, M, Willis, A, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How Tyrosine 15 Phosphorylation Inhibits the Activity of Cyclin-Dependent Kinase 2-Cyclin A.
J.Biol.Chem., 282, 2007
|
|
4BCQ
 
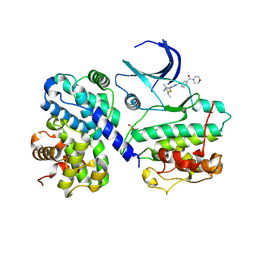 | | Structure of CDK2 in complex with cyclin A and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]-2-{[3-(morpholin-4-ylcarbonyl)phenyl]amino}pyrimidine-5-carbonitrile, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
2C6O
 
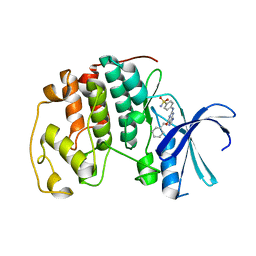 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2CCH
 
 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
2C68
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDROPYRAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C4G
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-533514 | | Descriptor: | (3Z)-5-ACETYL-3-(BENZOYLIMINO)-3,6-DIHYDROPYRROLO[3,4-C]PYRAZOL-5-IUM, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Fancelli, D, Vulpetti, A, Amici, R, Villa, M, Pittala, V, Ciomei, M, Mercurio, C, Bischoff, J.R, Roletto, F, Varasi, M, Brasca, M.G. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3-Amino-1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: A New Class of Cdk2 Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C5X
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, Mcinnes, C, Pandalaneni, S.R, Mcnae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2BHE
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 5-BROMO-INDIRUBINE | | Descriptor: | (2Z)-5'-BROMO-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE AMMONIATE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-10 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
5EIF
 
 | |
5EHG
 
 | |
4JQ3
 
 | | AKR1C2 complex with zomepirac | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, L(+)-TARTARIC ACID, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
4BCW
 
 | | Carbonic anhydrase IX mimic in complex with (E)-2-(5-bromo-2- hydroxyphenyl)ethenesulfonic acid | | Descriptor: | (E)-2-(5-BROMO-2-HYDROXYPHENYL)ETHENESULFONIC ACID, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfocoumarins (1,2-Benzoxathiine-2,2-Dioxides): A Class of Potent and Isoform-Selective Inhibitors of Tumor-Associated Carbonic Anhydrases.
J.Med.Chem., 56, 2013
|
|
4L1W
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
