2X7U
 
 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | Descriptor: | (9BETA,14BETA,17BETA)-17-HYDROXY-2-METHOXYESTRA-1,3,5(10)-TRIEN-3-YL SULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Non-Steroidal Inhibitors.
Biochemistry, 49, 2010
|
|
2X1N
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | 2-METHYL-N-[(1Z)-3-NITROCYCLOHEXA-2,4-DIEN-1-YLIDENE]-4,5-DIHYDRO[1,3]THIAZOLO[4,5-H]QUINAZOLIN-8-AMINE, ACE-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M, McIntyre, N.A, Griffiths, G, Barnett, A.L, Slawin, A.M.Z, Jackson, W, Thomas, M, Zheleva, D.I, Wang, S, Blake, D.G, Westwood, N.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
2W1H
 
 | | Fragment-Based Discovery of the Pyrazol-4-yl urea (AT9283), a Multi- targeted Kinase Inhibitor with Potent Aurora Kinase Activity | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[3-(1H-BENZIMIDAZOL-2-YL)-1H-PYRAZOL-4-YL]BENZAMIDE | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
1LBK
 
 | | Crystal structure of a recombinant glutathione transferase, created by replacing the last seven residues of each subunit of the human class pi isoenzyme with the additional C-terminal helix of human class alpha isoenzyme | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase class pi chimaera (CODA), ... | | Authors: | Kong, G.K.W, Micaloni, C, Mazzetti, A.P, Nuccetelli, M, Antonini, G, Stella, L, McKinstry, W.J, Polekhina, G, Rossjohn, J, Federici, G, Ricci, G, Parker, M.W, Lo Bello, M. | | Deposit date: | 2002-04-04 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering a new C-terminal tail in the H-site of human glutathione transferase P1-1: structural and functional consequences.
J.Mol.Biol., 325, 2003
|
|
2WIP
 
 | | STRUCTURE OF CDK2-CYCLIN A COMPLEXED WITH 8-ANILINO-1-METHYL-4,5-DIHYDRO- 1H-PYRAZOLO[4,3-H] QUINAZOLINE-3-CARBOXYLIC ACID | | Descriptor: | 1-methyl-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxylic acid, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
4A9E
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 3-methyl-1,2,3,4- tetrahydroquinazolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, BROMODOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 1: Inhibitor Binding Modes and Implications for Lead Discovery.
J.Med.Chem., 55, 2012
|
|
4ALH
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 3,5 dimethyl-4-phenyl-1,2- oxazole | | Descriptor: | 1,2-ETHANEDIOL, 3,5 DIMETHYL-4-PHENYL-1,2-OXAZOLE, BROMODOMAIN CONTAINING 2, ... | | Authors: | Chung, C.W, Bamborough, P. | | Deposit date: | 2012-03-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 2: Optimization of Phenylisoxazole Sulfonamides.
J.Med.Chem., 55, 2012
|
|
4JQ1
 
 | | AKR1C2 complex with naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
2XU3
 
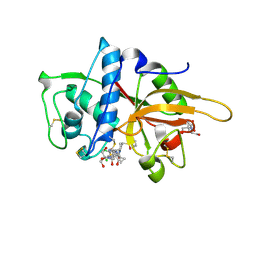 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-1-[1-(5-chlorothiophen-2-yl)cyclopropyl]carbonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATHEPSIN L1, ... | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6Q50
 
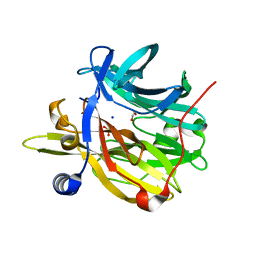 | | Structure of MPT-4, a mannose phosphorylase from Leishmania mexicana, in complex with phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, MPT-4, PHOSPHATE ION, ... | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
5DQN
 
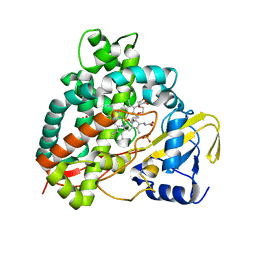 | | Polyethylene 600-bound form of P450 CYP125A3 mutant from Myobacterium Smegmatis - W83Y | | Descriptor: | CITRIC ACID, Cytochrome P450 CYP125, PENTAETHYLENE GLYCOL, ... | | Authors: | Ortiz de Montellano, P.J, Frank, D.J, Waddling, C.A. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis.
Biochemistry, 54, 2015
|
|
8SJX
 
 | |
6R1H
 
 | |
8SJY
 
 | |
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
5K2G
 
 | |
8ROX
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
2Y32
 
 | | Crystal structure of bradavidin | | Descriptor: | BLR5658 PROTEIN | | Authors: | Leppiniemi, J, Gronroos, T, Johnson, M.S, Kulomaa, M.S, Hytonen, V.P, Airenne, T.T. | | Deposit date: | 2010-12-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Bradavidin - C-Terminal Residues Act as Intrinsic Ligands.
Plos One, 7, 2012
|
|
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 2024
|
|
8RQH
 
 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
2C5O
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2C5Y
 
 | | DIFFERENTIAL BINDING OF INHIBITORS TO ACTIVE AND INACTIVE CDK2 PROVIDES INSIGHTS FOR DRUG DESIGN | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2XU5
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-(4-methylphenyl)cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, CITRATE ANION, ... | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2BKZ
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-404611 | | Descriptor: | 1-[4-(AMINOSULFONYL)PHENYL]-1,6-DIHYDROPYRAZOLO[3,4-E]INDAZOLE-3-CARBOXAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | DAlessio, R, Bargiottia, A, Metz, S, Brasca, M.G, Cameron, A, Ermoli, A, Marsiglio, A, Polucci, P, Roletto, F, Tibolla, M, Vazquez, M.L, Vulpetti, A, Pevarello, P. | | Deposit date: | 2005-02-23 | | Release date: | 2006-03-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Benzodipyrazoles: A New Class of Potent Cdk2 Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BHH
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 4-HYDROXYPIPERINDINESULFONYL-INDIRUBINE | | Descriptor: | (2E,3S)-3-HYDROXY-5'-[(4-HYDROXYPIPERIDIN-1-YL)SULFONYL]-3-METHYL-1,3-DIHYDRO-2,3'-BIINDOL-2'(1'H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
