3O95
 
 | | Crystal Structure of Human DPP4 Bound to TAK-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a 3-Pyridylacetic Acid Derivative (TAK-100) as a Potent, Selective and Orally Active Dipeptidyl Peptidase IV (DPP-4) Inhibitor.
J.Med.Chem., 53, 2011
|
|
8D7M
 
 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
8D7O
 
 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
8D7P
 
 | | Human Casein kinase 1 delta in complex with phosphorylated Drosophila PERIOD peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
8D7N
 
 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
2YCL
 
 | | complete structure of the corrinoid,iron-sulfur protein including the N-terminal domain with a 4Fe-4S cluster | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE CORRINOID/IRON-SULFUR PROTEIN, GAMMA SUBUNIT, CO DEHYDROGENASE/ACETYL-COA SYNTHASE, ... | | Authors: | Goetzl, S, Jeoung, J.H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
3B0H
 
 | | Assimilatory nitrite reductase (Nii4) from tobbaco root | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B2I
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3BIX
 
 | | Crystal structure of the extracellular esterase domain of Neuroligin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
3B0J
 
 | | M175E mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B2J
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3B6Q
 
 | |
3BFU
 
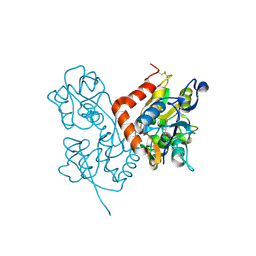 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
2YCJ
 
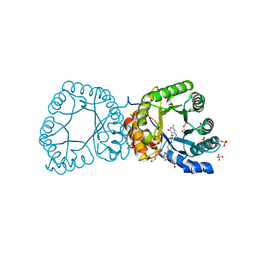 | | methyltransferase bound with methyltetrahydrofolate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-METHYLTETRAHYDROFOLATE CORRINOID/IRON SULFUR PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Goetzl, S, Jeoung, J.-H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
4NXP
 
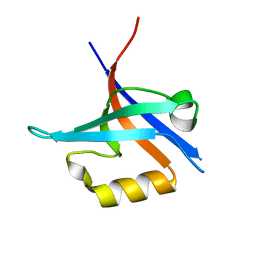 | | Crystal Structure of Free T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
3B6W
 
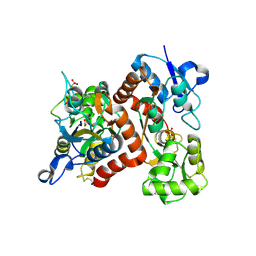 | |
8DGO
 
 | |
3BOK
 
 | |
3OPM
 
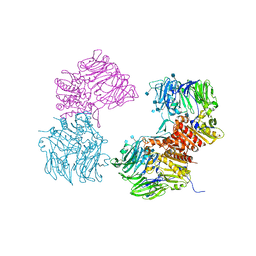 | | Crystal Structure of Human DPP4 Bound to TAK-294 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(aminomethyl)-2-(2-methylpropyl)-1-oxo-4-phenyl-1,2-dihydroisoquinolin-6-yl]oxy}acetamide, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-09-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of 3-aminomethyl-1,2-dihydro-4-phenyl-1-isoquinolones: a new class of potent, selective, and orally active non-peptide dipeptidyl peptidase IV inhibitors that form a unique interaction with Lys554.
Bioorg.Med.Chem., 19, 2011
|
|
2YCI
 
 | | methyltransferase native | | Descriptor: | 5-METHYLTETRAHYDROFOLATE CORRINOID/IRON SULFUR PROTEIN METHYLTRANSFERASE, SULFATE ION | | Authors: | Goetzl, S, Jeoung, J.H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
3BWJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-(6-{[(1R)-4-carbamimidamido-1-{[(1R)-4-carbamimidamido-1-carbamoylbutyl]carbamoyl}butyl]amino}-6-oxohexyl)-3,4-dihydroxytetrahydrofuran-2-carboxamide, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Lavogina, D, Koenig, N, Uri, A, Bossemeyer, D. | | Deposit date: | 2008-01-09 | | Release date: | 2009-02-03 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases.
J.Med.Chem., 52, 2009
|
|
7XS7
 
 | | structure of a membrane-integrated glycosyltransferase | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, Chitin synthase 1, DODECANE, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
7XS6
 
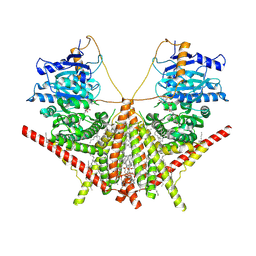 | | structure of a membrane-integrated glycosyltransferase with inhibitor | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
3C9L
 
 | |
