4ZE0
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140F mutant complexed with Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
8GGX
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #16 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGY
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #17 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGW
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #15 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
4ZKA
 
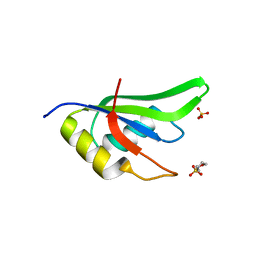 | | High Resolution Crystal Structure of Fox1 RRM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RNA binding protein fox-1 homolog 1, SULFATE ION, ... | | Authors: | Blatter, M, Allain, F.H.-T. | | Deposit date: | 2015-04-30 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Crystal Structure of Fox1 RRM
To Be Published
|
|
8GGZ
 
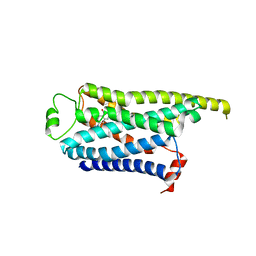 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #18 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
4ZKN
 
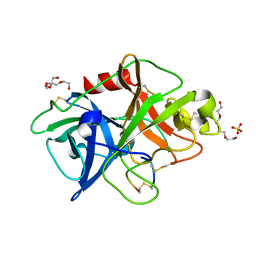 | |
5D0J
 
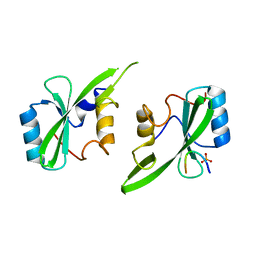 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
8GH0
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #19 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GH1
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #20 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5CYT
 
 | | REFINEMENT OF MYOGLOBIN AND CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Takano, T. | | Deposit date: | 1988-01-14 | | Release date: | 1989-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refinement of Myoglobin and Cytochrome C
Methods and Applications in Crystallographic Computing, 1984
|
|
6SEX
 
 | | X-ray structure of the gold/lysozyme adduct formed upon 21h exposure of protein crystals to compound 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraro, G, Giorgio, A, Merlino, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
4ZFB
 
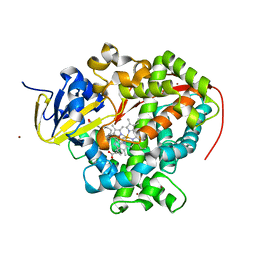 | | Cytochrome P450 pentamutant from BM3 bound to Palmitic Acid | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
5CYX
 
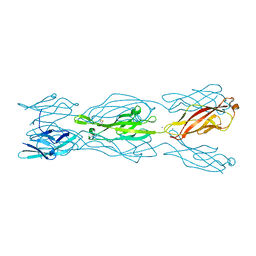 | |
4Q5P
 
 | | Lysine-Ligated Yeast Iso-1 Cytochrome C | | Descriptor: | Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Amacher, J.F, Zhu, M.Q, Zhong, F, Pletneva, E.V, Madden, D.R. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Compact Structure of Cytochrome c Trapped in a Lysine-Ligated State: Loop Refolding and Functional Implications of a Conformational Switch.
J.Am.Chem.Soc., 137, 2015
|
|
4QP3
 
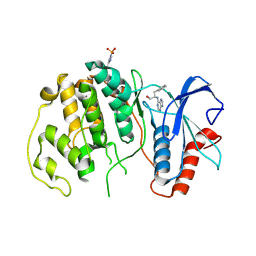 | |
7UAH
 
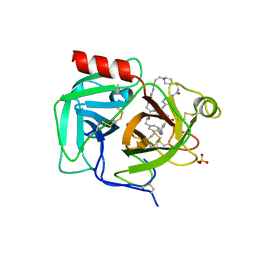 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7ULY
 
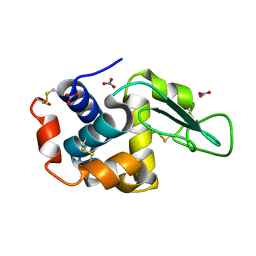 | | MicroED structure of triclinic lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Gonen, T. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.87 Å) | | Cite: | Hydrogens and hydrogen-bond networks in macromolecular MicroED data.
J Struct Biol X, 6, 2022
|
|
8DYZ
 
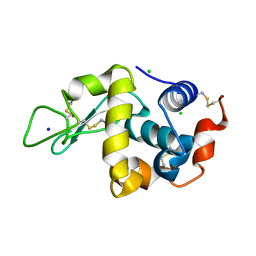 | |
5YMX
 
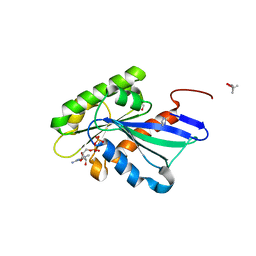 | | Myxococcus xanthus MglA in GDP bound conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Baranwal, J, Gayathri, P. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Allosteric regulation of a prokaryotic small Ras-like GTPase contributes to cell polarity oscillations in bacterial motility.
Plos Biol., 17, 2019
|
|
6BX6
 
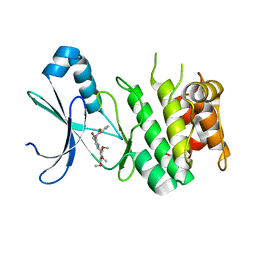 | | AMP-Activated protein kinase (AMPK) inhibition by SBI-0206965: alpha 2 kinase domain bound to SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Dite, T.A, Langendorf, C.G, Scott, J.W, Oakhill, J.S. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965.
J. Biol. Chem., 293, 2018
|
|
6SNK
 
 | | Crystal structure of the Collagen VI alpha3 N2 domain | | Descriptor: | Collagen alpha-3(VI) chain | | Authors: | Gebauer, J.M, Degefa, H.S, Paulsson, M, Wagener, R, Baumann, U. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a collagen VI alpha 3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations.
J.Biol.Chem., 295, 2020
|
|
8GGG
 
 | | RNase A-Adenosine 5'-Hexaphosphate (RNaseA.p6A) | | Descriptor: | GLYCEROL, Ribonuclease pancreatic, adenosine 5'-hexaphosphate | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A.
Acs Cent.Sci., 10, 2024
|
|
6SO1
 
 | | Fragment N13569a in complex with MAP kinase p38-alpha | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-[[(2~{R})-oxolan-2-yl]methyl]methanamine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
4Z50
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20D25N with Tucked Flap | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Agniswamy, J, Shen, C.-H, Weber, I.T. | | Deposit date: | 2015-04-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational variation of an extreme drug resistant mutant of HIV protease.
J.Mol.Graph.Model., 62, 2015
|
|
