3SQI
 
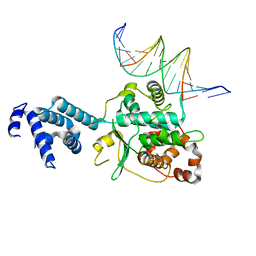 | | DNA binding domain of Ndc10 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*AP*AP*TP*T)-3'), KLLA0E03807p | | Authors: | Cho, U.S, Harrison, S.C. | | Deposit date: | 2011-07-05 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8245 Å) | | Cite: | Ndc10 is a platform for inner kinetochore assembly in budding yeast.
Nat.Struct.Mol.Biol., 19, 2011
|
|
2BRF
 
 | |
2K6G
 
 | |
3L1Z
 
 | |
2MRF
 
 | | NMR structure of the ubiquitin-binding zinc finger (UBZ) domain from human Rad18 | | Descriptor: | E3 ubiquitin-protein ligase RAD18, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
2IQC
 
 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
8R64
 
 | |
3ZQS
 
 | | Human FANCL central domain | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE FANCL, HEXAETHYLENE GLYCOL, PROLINE, ... | | Authors: | Hodson, C, Cole, A.R, Purkiss-Trew, A, Walden, H. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Fancl, the E3 Ligase in the Fanconi Anemia Pathway.
J.Biol.Chem., 286, 2011
|
|
3U7E
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6CHS
 
 | | Cdc48-Npl4 complex in the presence of ATP-gamma-S | | Descriptor: | MAGNESIUM ION, Npl4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kim, K.H, Bodnar, N.O, Walz, T, Rapoport, T.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8ITY
 
 | | human RNA polymerase III pre-initiation complex closed DNA 1 | | Descriptor: | DNA (82-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
6GYM
 
 | | Structure of a yeast closed complex with distorted DNA (CCdist) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Dienemann, C, Schwalb, B, Schilbach, S, Cramer, P. | | Deposit date: | 2018-06-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Promoter Distortion and Opening in the RNA Polymerase II Cleft.
Mol. Cell, 73, 2019
|
|
4R8P
 
 | | Crystal structure of the Ring1B/Bmi1/UbcH5c PRC1 ubiquitylation module bound to the nucleosome core particle | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase RING2, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | McGinty, R.K, Henrici, R.C, Tan, S. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2846 Å) | | Cite: | Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome.
Nature, 514, 2014
|
|
3E0C
 
 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | Descriptor: | DNA damage-binding protein 1 | | Authors: | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
5CY1
 
 | |
2CRX
 
 | |
8X7I
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7K
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | Descriptor: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7J
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy | | Descriptor: | DNA (144-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
