3LFE
 
 | | Human p38 MAP Kinase in Complex with RL116 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{4-[2-(pyridin-4-ylmethoxy)ethyl]-1,3-thiazol-2-yl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LKW
 
 | | Crystal Structure of Dengue Virus 1 NS2B/NS3 protease active site mutant | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chandramouli, S, Joseph, J.S, Daudenarde, S, Gatchalian, J, Cornillez-Ty, C, Kuhn, P. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family.
J.Virol., 84, 2010
|
|
3LGV
 
 | | H198P mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LIB
 
 | |
3LKS
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1980 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LOG
 
 | | Crystal structure of MbtI from Mycobacterium tuberculosis | | Descriptor: | CARBONATE ION, GLYCEROL, Isochorismate synthase/isochorismate-pyruvate lyase mbtI, ... | | Authors: | Bulloch, E.M.M, Lott, J.S, Baker, E.N, Johnston, J.M. | | Deposit date: | 2010-02-03 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI).
Chemmedchem, 5, 2010
|
|
3LIS
 
 | |
3LJI
 
 | | CRYSTAL STRUCTURE OF putative geranyltranstransferase from Pseudomonas fluorescens Pf-5 | | Descriptor: | Geranyltranstransferase | | Authors: | Malashkevich, V.N, Toro, R, Patskovsky, Y, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | CRYSTAL STRUCTURE OF putative geranyltranstransferase from Pseudomonas fluorescens Pf-5
To be Published
|
|
3LL9
 
 | |
3LFD
 
 | | Human p38 MAP Kinase in Complex with RL113 | | Descriptor: | 1-{4-[2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LMV
 
 | | D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-tyrosyl-tRNA(Tyr) deacylase, SULFITE ION | | Authors: | Manickam, Y, Khan, S, Bhatt, T.K, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LH5
 
 | |
3LI8
 
 | |
3LPF
 
 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 1-((6,7-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(3-methoxyphenyl)thiourea | | Descriptor: | 1-[(6,7-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(3-methoxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Alleviating cancer drug toxicity by inhibiting a bacterial enzyme.
Science, 330, 2010
|
|
3LKT
 
 | |
3LR3
 
 | |
3LOZ
 
 | |
3LUZ
 
 | |
3L6A
 
 | | Crystal structure of the C-terminal region of Human p97 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Eukaryotic translation initiation factor 4 gamma 2, ... | | Authors: | Fan, S. | | Deposit date: | 2009-12-23 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal region of human p97/DAP5.
Proteins, 78, 2010
|
|
3L5H
 
 | | Crystal structure of the full ectodomain of human gp130: New insights into the molecular assembly of receptor complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6 receptor subunit beta, SULFATE ION, ... | | Authors: | Xu, Y, Garrett, T.P.J, Zhang, J.G. | | Deposit date: | 2009-12-21 | | Release date: | 2010-05-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
3L5V
 
 | |
3L6Q
 
 | | Crystal structure of the N-terminal domain of HSP70 from Cryptosporidium parvum (CGD2_20) | | Descriptor: | Heat shock 70 (HSP70) protein, MAGNESIUM ION, SULFATE ION | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Lew, J, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of the N-terminal domain of HSP70 from Cryptosporidium parvum (CGD2_20)
To be Published
|
|
3L7Z
 
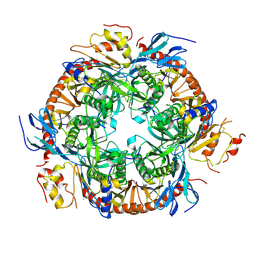 | | Crystal structure of the S. solfataricus archaeal exosome | | Descriptor: | Probable exosome complex RNA-binding protein 1, Probable exosome complex exonuclease 1, Probable exosome complex exonuclease 2, ... | | Authors: | Lu, C, Ding, F, Ke, A. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring.
Plos One, 5, 2010
|
|
3LR0
 
 | |
3LEO
 
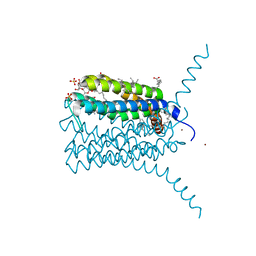 | | Structure of human Leukotriene C4 synthase mutant R31Q in complex with glutathione | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Niegowski, D, Martinez-Molina, D, Rinaldo-Matthis, A, Nordlund, P, Haeggstrom, J. | | Deposit date: | 2010-01-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arginine 104 is a key catalytic residue in leukotriene C4 synthase.
J.Biol.Chem., 285, 2010
|
|
