8ZRT
 
 | | Cryo-EM structure focused on the receptor of the ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1 | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of endothelin ETB receptor-Gi complex in a conformation stabilized by the unique NPxxL motif
Commun Biol, 2024
|
|
9B71
 
 | | Cryo-EM structure of MraY in complex with analogue 3 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]-2-[[4-[[[(2~{S})-5-carbamimidamido-2-(hexadecanoylamino)pentanoyl]amino]methyl]phenyl]methylamino]propanoic acid, MraYAA Nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
9IYE
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution.
To Be Published
|
|
9BCM
 
 | |
9FJY
 
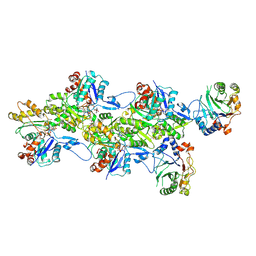 | | Structure of the DNase I- and phalloidin-bound pointed end of F-actin (conformer 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human dopamine transporter in complex with cocaine.
Nature, 632, 2024
|
|
8ZFL
 
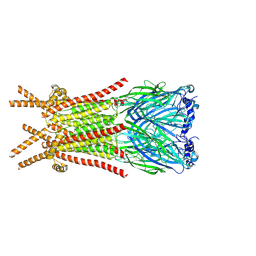 | | Caenorhabditis elegans ACR-23 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betaine receptor acr-23,Soluble cytochrome b562 | | Authors: | Chen, Q.F, Liu, F.L, Li, T.Y, Gong, H.H, Guo, F, Liu, S. | | Deposit date: | 2024-05-08 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of acetylcholine receptor like-23 (ACR-23) activation by anthelmintics monepantel and betaine
To Be Published
|
|
9IHS
 
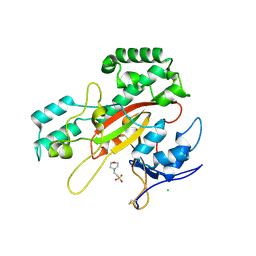 | | Microbial transglutaminase mutant - D3C/G283C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suzuki, M, Date, M, Kashiwagi, T, Takahashi, K, Nakamura, A, Tanokura, M, Suzuki, E, Yokoyama, K. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Random mutagenesis and disulfide bond formation improved thermostability in microbial transglutaminase.
Appl.Microbiol.Biotechnol., 2024
|
|
9AVT
 
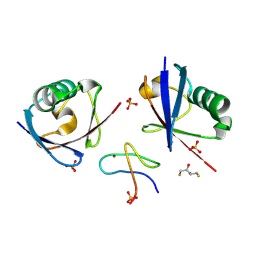 | | Structure of TAB2 NZF domain bound to K6 / Lys6-linked diubiquitin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TGF-beta-activated kinase 1 and MAP3K7-binding protein 2, ... | | Authors: | Michel, M.A, Scutts, S, Komander, D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of TAB2 NZF domain bound to K6 / Lys6-linked diubiquitin
To be published
|
|
9F63
 
 | |
9AYC
 
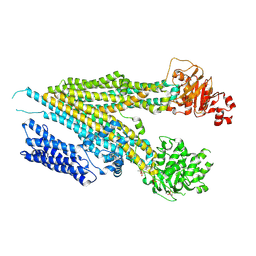 | |
8ZYO
 
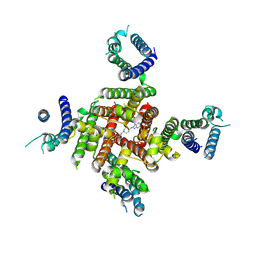 | | Cryo-EM Structure of astemizole-bound hERG Channel | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
9C62
 
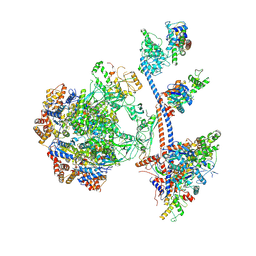 | | P400 subcomplex of the native human TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (5.28 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9FJM
 
 | | Cryo-EM structure of the phalloidin-bound pointed end of the actin filament. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9AYX
 
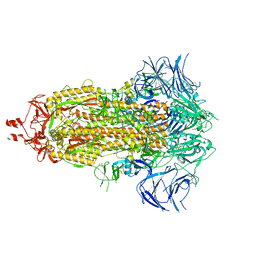 | |
9EQH
 
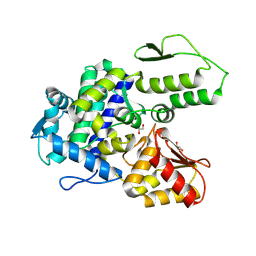 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the inhibitor space of the WWP1 and WWP2 HECT E3 ligases.
J Enzyme Inhib Med Chem, 39, 2024
|
|
9FMM
 
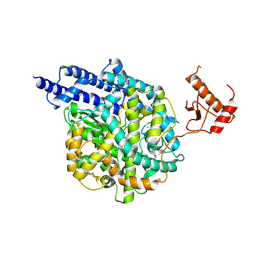 | | Structure of human ACE2 in complex with a fluorinated small molecule inhibitor | | Descriptor: | (2~{S})-2-[[(2~{S})-3-[3-[(3-chloranyl-5-fluoranyl-phenyl)methyl]imidazol-4-yl]-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-4-methyl-pentanoic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benoit, R.B, Rodrigues, M.J, Wieser, M.M. | | Deposit date: | 2024-06-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of radiofluorinated MLN-4760 derivatives for PET imaging of the SARS-CoV-2 entry receptor ACE2.
Eur J Nucl Med Mol Imaging, 2024
|
|
9IK1
 
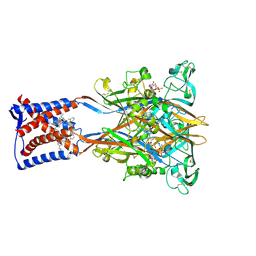 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
9C4B
 
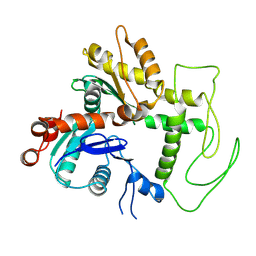 | | Second BAF53a of the human TIP60 complex | | Descriptor: | Actin-like protein 6A | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
8ZMR
 
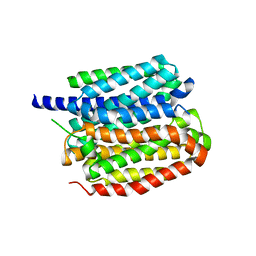 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
9ATP
 
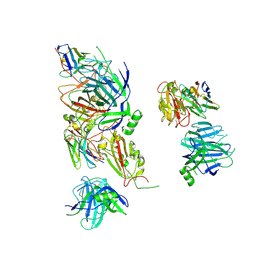 | |
9CTH
 
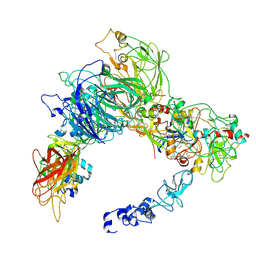 | | Preliminary map of the Prothrombin-prothrombinase complex on nano discs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated Factor V (FVa) heavy chain, Activated Factor V (FVa) light chain, ... | | Authors: | Stojanovski, B.M, Mohammed, B.M, Di Cera, E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | The Prothrombin-Prothrombinase Interaction.
Subcell Biochem, 104, 2024
|
|
9AX8
 
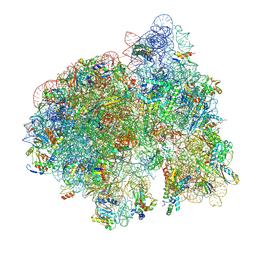 | | 70S initiation complex (tRNA-fMet M1, initiation factor 2 + CUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 2024
|
|
9AYY
 
 | |
9C6N
 
 | | ARP module of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
