8B6X
 
 | |
6HAT
 
 | | Globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, ZINC ION, mRNA export factor ICP27 homolog | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
8E7N
 
 | | Crystal structure of beluga whale Gammacoronavirus SW1 Mpro with GC-376 captured in two conformational states | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, GLYCEROL, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Inhibitor-Bound Main Protease from Delta- and Gamma-Coronaviruses.
Viruses, 15, 2023
|
|
4RC6
 
 | | Crystal structure of cyanobacterial aldehyde-deformylating oxygenase 122F mutant | | Descriptor: | Aldehyde decarbonylase, FE (II) ION | | Authors: | Jia, C.J, Li, M, Li, J.J, Zhang, J.J, Zhang, H.M, Cao, P, Pan, X.W, Lu, X.F, Chang, W.R. | | Deposit date: | 2014-09-14 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases.
Protein Cell, 6, 2015
|
|
4RTH
 
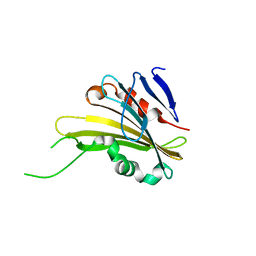 | | The crystal structure of PsbP from Zea mays | | Descriptor: | Membrane-extrinsic protein of photosystem II PsbP | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RTI
 
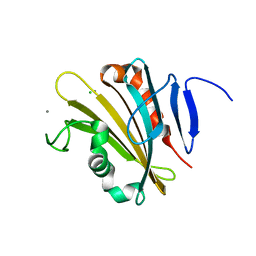 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
6LXT
 
 | | Structure of post fusion core of 2019-nCoV S2 subunit | | Descriptor: | Spike protein S2, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Zhu, Y, Sun, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion.
Cell Res., 30, 2020
|
|
3RAE
 
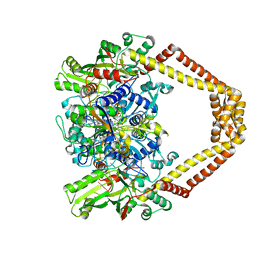 | | Quinolone(Levofloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3RAD
 
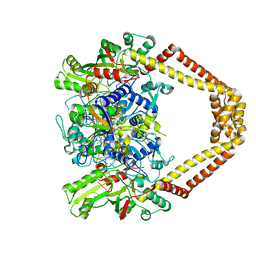 | | Quinolone(Clinafloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', 5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*G)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-04-25 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
6OAF
 
 | | Sudan virus nucleoprotein core domain in complex with VP35 chaperoning peptide | | Descriptor: | Polymerase cofactor VP35,Nucleoprotein | | Authors: | Landeras-Bueno, S, Oda, S, Norris, M.J, Li Salie, Z, Ollmann Saphire, E. | | Deposit date: | 2019-03-15 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sudan Ebolavirus VP35-NP Crystal Structure Reveals a Potential Target for Pan-Filovirus Treatment.
Mbio, 10, 2019
|
|
6HE7
 
 | | 20S proteasome from Archaeoglobus fulgidus | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5KVT
 
 | | THE STRUCTURE OF TRKA KINASE DOMAIN BOUND TO THE INHIBITOR ENTRECTINIB | | Descriptor: | Entrectinib, GLYCEROL, High affinity nerve growth factor receptor | | Authors: | Jin, L, Yan, S, Wei, G, Li, G, Harris, J, Vernier, J.-M. | | Deposit date: | 2016-07-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antitumor Activity and Safety of the Pan-TRK, ROS1, and ALK inhibitor Entrectinib (RXDX-101): Combined Results from Two Phase I Trials
To Be Published
|
|
7MYJ
 
 | | Structure of full length human AMPK (a2b1g1) in complex with a small molecule activator MSG011 | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Ovens, A.J, Gee, Y.S, Ling, N.X.Y, Waters, N.J, Yu, D, Scott, J.W, Parker, M.W, Hoffman, N.J, Kemp, B.E, Baell, J.B, Oakhill, J.S, Langendorf, C.G. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-function analysis of the AMPK activator SC4 and identification of a potent pan AMPK activator.
Biochem.J., 479, 2022
|
|
8U37
 
 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with NVP-CJL037 at 2.48-A resolution | | Descriptor: | (6M)-3-amino-N-{4-[(3R,4S)-4-amino-3-methoxypiperidin-1-yl]pyridin-3-yl}-6-[3-(trifluoromethoxy)pyridin-2-yl]pyrazine-2-carboxamide, MAGNESIUM ION, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8UAK
 
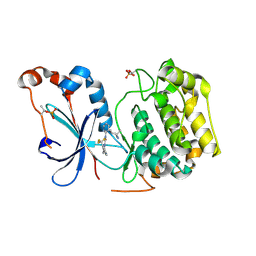 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8A4R
 
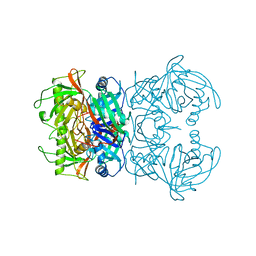 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic orthorhombic data at 3.59 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
8A3F
 
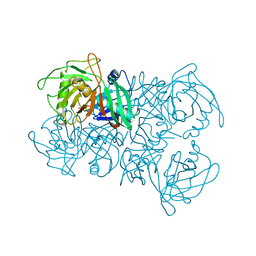 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic tetragonal data at 3.15 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
6WGX
 
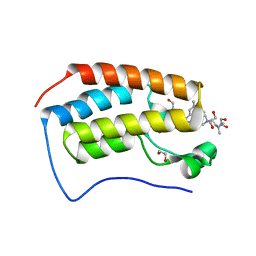 | | Cocrystal of BRD4(D1) with a selective inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(1-{1-[2-(dimethylamino)ethyl]piperidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-5-yl)-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | Descriptor: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
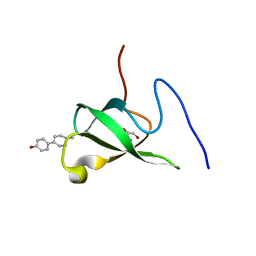 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
4I3H
 
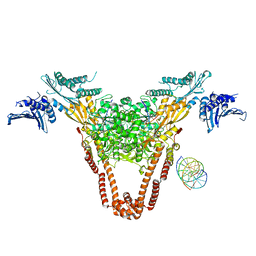 | | A three-gate structure of topoisomerase IV from Streptococcus pneumoniae | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*GP*CP*GP*GP*TP*AP*AP*TP*AP*CP*GP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*AP*AP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*TP*TP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*CP*GP*TP*AP*TP*TP*AP*CP*CP*GP*CP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Crevel, I, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2012-11-26 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.
Nucleic Acids Res., 41, 2013
|
|
4JUO
 
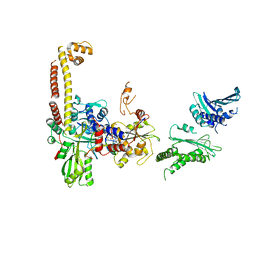 | | A low-resolution three-gate structure of topoisomerase IV from Streptococcus pneumoniae in space group H32 | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA topoisomerase 4 subunit A, DNA topoisomerase 4 subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Crevel, I, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.53 Å) | | Cite: | Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.
Nucleic Acids Res., 41, 2013
|
|
2KJX
 
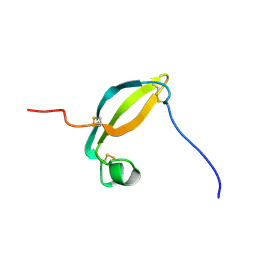 | | Solution structure of the extracellular domain of JTB | | Descriptor: | Jumping translocation breakpoint protein | | Authors: | Rousseau, F, Lingel, A, Pan, B, Fairbrother, W.J, Bazan, F. | | Deposit date: | 2009-06-10 | | Release date: | 2010-08-11 | | Last modified: | 2012-04-18 | | Method: | SOLUTION NMR | | Cite: | The structure of the extracellular domain of the jumping translocation breakpoint protein reveals a variation of the midkine fold.
J.Mol.Biol., 415, 2012
|
|
2HDE
 
 | |
