6BQ8
 
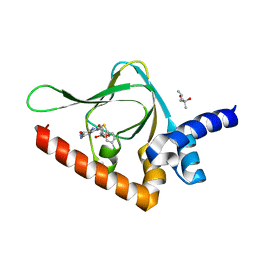 | | Joint X-ray/neutron structure of PKG II CNB-B domain in complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(~2~H_2_)amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2-hydroxy-7-(~2~H)hydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl](~2~H)-1,9-dihydro-6H-purin-6-one, STRONTIUM ION, ... | | Authors: | Kim, C, Kovalevsky, A, Gerlits, O. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallography Detects Differences in Protein Dynamics: Structure of the PKG II Cyclic Nucleotide Binding Domain in Complex with an Activator.
Biochemistry, 57, 2018
|
|
5ZJ3
 
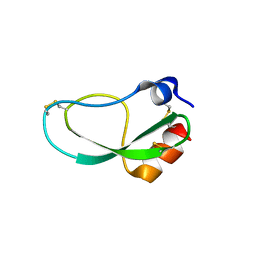 | |
8GYE
 
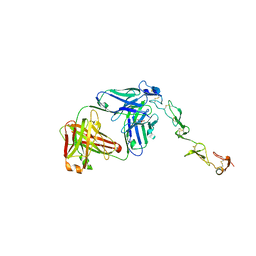 | |
4OIK
 
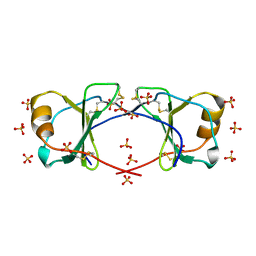 | | (Quasi-)Racemic X-ray crystal structure of glycosylated chemokine Ser-CCL1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1, CITRIC ACID, ... | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8GEY
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with 4-(hydroxymethyl)-1-[(4-methoxy-5,6,7,8-tetrahydronaphthalen-1-yl)sulfonyl]piperidin-4-ol | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(hydroxymethyl)-1-(4-methoxy-5,6,7,8-tetrahydronaphthalene-1-sulfonyl)piperidin-4-ol, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
5VZN
 
 | | Wild-type sperm whale myoglobin with nitric oxide | | Descriptor: | GLYCEROL, Myoglobin, NITRIC OXIDE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
8GD2
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-(2-thienylmethyl)methanamine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(thiophen-2-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
5VIZ
 
 | |
4ORW
 
 | | Three-dimensional structure of the C65A-K59A double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OS8
 
 | | Three-dimensional structure of the C65A-W54F-W112F triple mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8GEV
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine | | Descriptor: | 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
4OO5
 
 | |
6E8F
 
 | |
8GEU
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with methyl({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)[(1-methylpyrazol-4-yl)methyl]amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(1-methyl-1H-pyrazol-4-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
5VD0
 
 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH MK1775 | | Descriptor: | 1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, DI(HYDROXYETHYL)ETHER, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
7UOS
 
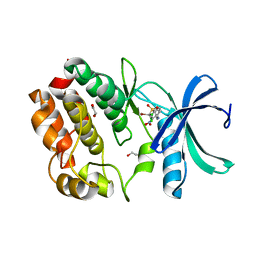 | | Structure of WNK1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 4-[bromo(dichloro)methanesulfonyl]-N-cyclohexyl-2-nitroaniline, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J, Akella, R. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of WNK1 inhibitor complex
To Be Published
|
|
6EAV
 
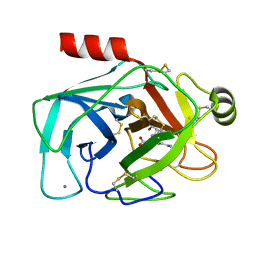 | |
6EB5
 
 | |
5VFJ
 
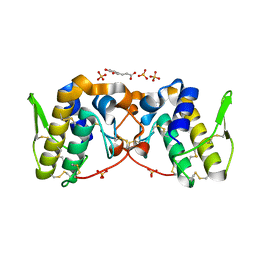 | |
7DIH
 
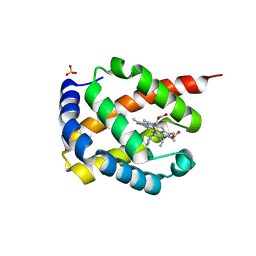 | |
6ER7
 
 | | CHEMOTAXIS PROTEIN CHEY FROM Pyrococcus horikoshiI | | Descriptor: | 120aa long hypothetical chemotaxis protein (CheY) | | Authors: | Paithankar, K.S, Enderle, M.E, Wirthensohn, D, Grininger, M, Oesterhelt, D. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the archaeal chemotaxis protein CheY in a domain-swapped dimeric conformation.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6ILJ
 
 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
5Z1C
 
 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
5VJ3
 
 | |
6I5K
 
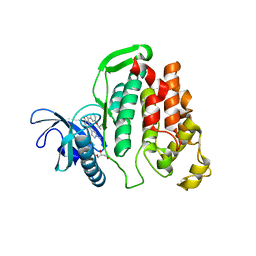 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN345 (derivative of compound 12h) | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-(3-propan-2-yloxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
