5TY6
 
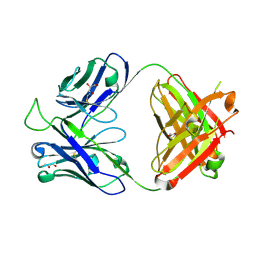 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 13-1b02 Fab. | | Descriptor: | GLYCEROL, VRC 315 13-1b02 Fab Heavy chain, VRC 315 13-1b02 Fab Light chain | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2016-11-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
3V0A
 
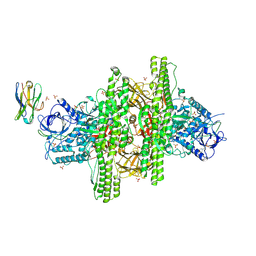 | | 2.7 angstrom crystal structure of BoNT/Ai in complex with NTNHA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BoNT/A, CALCIUM ION, ... | | Authors: | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
7U0Y
 
 | |
7ORB
 
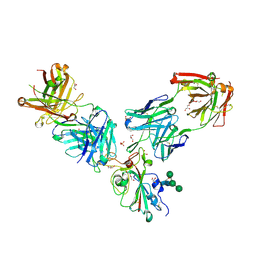 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7OR9
 
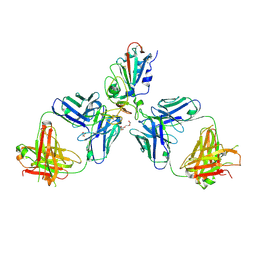 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7ORA
 
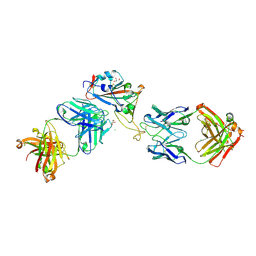 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7JKM
 
 | |
5EII
 
 | |
7KGV
 
 | | Crystal structure of sodium-coupled neutral amino acid transporter SLC38A9 in the N-terminal plugged form | | Descriptor: | Monoclonal antibody Fab heavy chain, Monoclonal antibody Fab light chain, Sodium-coupled neutral amino acid transporter 9 | | Authors: | Lei, H, Mu, X, Hattne, J, Gonen, T. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conformational change in the N terminus of SLC38A9 signals mTORC1 activation.
Structure, 29, 2021
|
|
2OBG
 
 | |
1F6A
 
 | | Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Wurzburg, B.A, Tarchevskaya, S.S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc (epsilon) RI (alpha).
Nature, 406, 2000
|
|
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
8GHK
 
 | |
7NEH
 
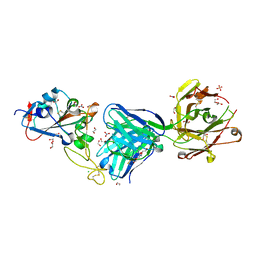 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-02-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
7WGT
 
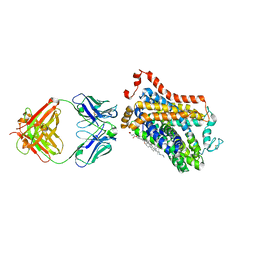 | | X-ray structure of thermostabilized Drosophila dopamine transporter with GABA transporter1-like substitutions in the binding site, in complex with NO711. | | Descriptor: | 1-(2-{[(diphenylmethylidene)amino]oxy}ethyl)-1,2,5,6-tetrahydropyridine-3-carboxylic acid, Antibody fragment (9D5) heavy chain, Antibody fragment (9D5) light chain, ... | | Authors: | Joseph, D, Penmatsa, A. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Embo J., 41, 2022
|
|
7CVT
 
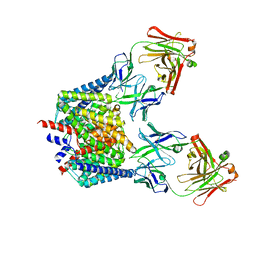 | | Crystal structure of the C85A/L194A/H234C mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
5NQX
 
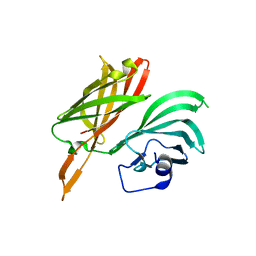 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 294. | | Descriptor: | Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein | | Authors: | Johnson, S, Jongerius, I, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQZ
 
 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
7CVS
 
 | | Crystal structure of the C85A/L194A mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
3SGE
 
 | | Crystal structure of mAb 17.2 in complex with R13 peptide | | Descriptor: | CALCIUM ION, Heavy Chain, Light Chain, ... | | Authors: | Pizarro, J.C, Boulot, G, Hontebeyrie, M, Bentley, G.A. | | Deposit date: | 2011-06-14 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex mAb 17.2 and the C-terminal region of Trypanosoma cruzi P2 Beta protein: implications in cross-reactivity
Plos Negl Trop Dis, 5, 2011
|
|
3C08
 
 | |
3QWQ
 
 | |
3QWR
 
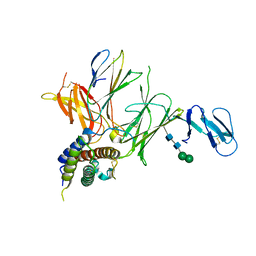 | | Crystal structure of IL-23 in complex with an adnectin | | Descriptor: | ADNECTIN, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Wei, A, Sheriff, S. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of adnectin/protein complexes reveal an expanded binding footprint.
Structure, 20, 2012
|
|
7K75
 
 | |
7K76
 
 | |
