3DYA
 
 | | HIV-1 RT with non-nucleoside inhibitor annulated Pyrazole 1 | | Descriptor: | 3-[6-bromo-2-fluoro-3-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-chlorobenzonitrile, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
1OQN
 
 | | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | | Descriptor: | Alzheimer's disease amyloid A4 protein homolog, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1 | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2003-03-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
4HGC
 
 | | Crystal structure of bovine trypsin complexed with sfti-1 analog containing a peptoid residue at position p1 | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Krzywda, S, Jaskolski, M, Rolka, K, Stawikowski, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of a proteolytically resistant analogue of (NLys)(5)SFTI-1 in complex with trypsin: evidence for the direct participation of the Ser214 carbonyl group in serine protease-mediated proteolysis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1L9Y
 
 | | FEZ-1-Y228A, A Mutant of the Metallo-beta-lactamase from Legionella gormanii | | Descriptor: | CHLORIDE ION, FEZ-1 b-lactamase, GLYCEROL, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Galleni, M, Dideberg, O. | | Deposit date: | 2002-03-27 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-beta-lactamase
from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.Mol.Biol., 325, 2003
|
|
3DZA
 
 | |
1G35
 
 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE IN COMPLEX WITH INHIBITOR, AHA024 | | Descriptor: | 2-[4-(HYDROXY-METHOXY-METHYL)-BENZYL]-7-(4-HYDROXYMETHYL-BENZYL)-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1LAMBDA6-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
1ZPA
 
 | | HIV Protease with Scripps AB-3 Inhibitor | | Descriptor: | Pol polyprotein, TERT-BUTYL 4-[({[1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL}AMINO)CARBONYL]BENZYLCARBAMATE | | Authors: | Brik, A, Alexandratos, J, Lin, Y.C, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors
Chembiochem, 6, 2005
|
|
3S8G
 
 | | 1.8 A structure of ba3 cytochrome c oxidase mutant (A120F) from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
2QVR
 
 | | E. coli Fructose-1,6-bisphosphatase: Citrate, Fru-2,6-P2, and Mg2+ bound | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CITRIC ACID, Fructose-1,6-bisphosphatase, ... | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of Mammalian and Bacterial Fructose-1,6-bisphosphatase Reveal the Basis for Synergism in AMP/Fructose 2,6-Bisphosphate Inhibition.
J.Biol.Chem., 282, 2007
|
|
4K3A
 
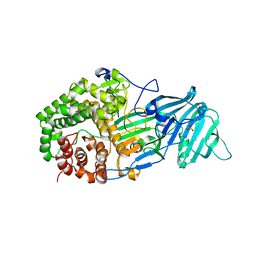 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
3C2H
 
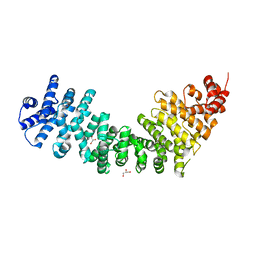 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
2QME
 
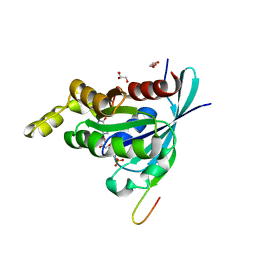 | | Crystal structure of human RAC3 in complex with CRIB domain of human p21-activated kinase 1 (PAK1) | | Descriptor: | CRIB domain of the Serine/threonine-protein kinase PAK 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J.M, Burgess-Brown, N, Bunkoczi, G, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 1 (PAK1).
To be Published
|
|
3E7A
 
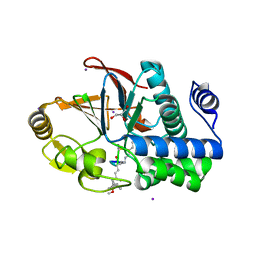 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin Nodularin-R | | Descriptor: | AZIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
2XIZ
 
 | | Protein kinase Pim-1 in complex with fragment-3 from crystallographic fragment screen | | Descriptor: | (E)-PYRIDIN-4-YL-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3EGG
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Spinophilin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3S8F
 
 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
3MLT
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a UG1033 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MQO
 
 | | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Stein, A.J, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A
To be Published
|
|
3BIK
 
 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2FLF
 
 | | Crystal structure of l-fuculose-1-phosphate aldolase from Thermus Thermophilus HB8 | | Descriptor: | fuculose-1-phosphate aldolase | | Authors: | Jeyakanthan, J, Yokoyama, S, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Purification, crystallization and preliminary X-ray crystallographic study of the L-fuculose-1-phosphate aldolase (FucA) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3S64
 
 | | Saposin-like protein Ac-SLP-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, Saposin-like protein 1 | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
2XJ1
 
 | | Protein kinase Pim-1 in complex with small molecule inibitor | | Descriptor: | (2E)-3-(3-{6-[(TRANS-4-AMINOCYCLOHEXYL)AMINO]PYRAZIN-2-YL}PHENYL)PROP-2-ENOIC ACID, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2AVQ
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
4D1F
 
 | |
