8I2J
 
 | |
8I2M
 
 | |
8I4I
 
 | |
4UCF
 
 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
4U6Z
 
 | |
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
5DOG
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-(Benzylamino)-3,5,6-trifluoro-4-[(2-phenylethyl)thio]benzene- sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-(benzylamino)-3,5,6-trifluoro-4-[(2-phenylethyl)sulfanyl]benzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrinsic Thermodynamics and Structures of 2,4- and 3,4-Substituted Fluorinated Benzenesulfonamides Binding to Carbonic Anhydrases.
ChemMedChem, 12, 2017
|
|
5H2U
 
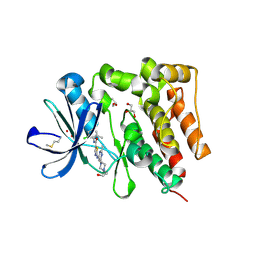 | | Crystal structure of PTK6 Kinase Domain complexed with Dasatinib | | Descriptor: | GLYCEROL, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
8SPN
 
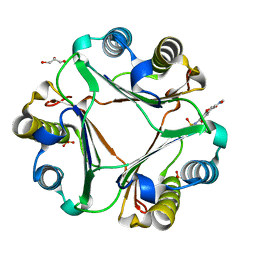 | | Crystal structure of macrophage migration inhibitory factor in complex with T614 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Iguratimod, ... | | Authors: | Pantouris, G, Lolis, E. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Iguratimod, an allosteric inhibitor of macrophage migration inhibitory factor (MIF), prevents mortality and oxidative stress in a murine model of acetaminophen overdose.
Mol Med, 30, 2024
|
|
8SON
 
 | |
5I3S
 
 | |
5DMU
 
 | | Structure of the NHEJ polymerase from Methanocella paludicola | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NHEJ Polymerase, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
2A8S
 
 | | 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear SnoRNA Decapping Nudix Hydrolase X29, Manganese and GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, U8 snoRNA-binding protein X29 | | Authors: | Scarsdale, J.N, Peculis, B.A, Wright, H.T. | | Deposit date: | 2005-07-08 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes
Structure, 14, 2006
|
|
2A8T
 
 | | 2.1 Angstrom Crystal Structure of the Complex Between the Nuclear U8 snoRNA Decapping Nudix Hydrolase X29, Manganese and m7G-PPP-A | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE, MANGANESE (II) ION, ... | | Authors: | Scarsdale, J.N, Peculis, B.A, Wright, H.T. | | Deposit date: | 2005-07-08 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes
Structure, 14, 2006
|
|
6T38
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
7KL0
 
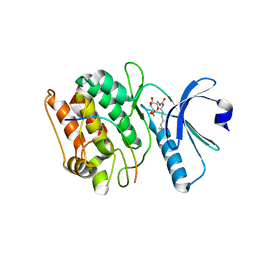 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
4LHI
 
 | |
5JNA
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and topiramate | | Descriptor: | ACETATE ION, Carbonic anhydrase 4, GLYCEROL, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5JQT
 
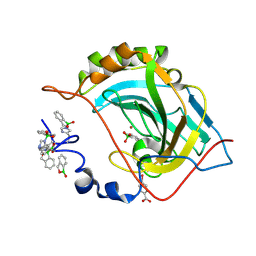 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH 7.4 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, 2,1-benzoxaborol-1(3H)-ol, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-05 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
7L03
 
 | |
7L4D
 
 | |
7L47
 
 | |
7L1H
 
 | |
2JDL
 
 | | Structure of C-terminal region of acidic P2 ribosomal protein complexed with trichosanthin | | Descriptor: | ACIDIC RIBOSOMAL PROTEIN P2, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Mak, A.N, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
