5S8L
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with E11289c (space group C2) | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | XChem group deposition
To Be Published
|
|
6SKW
 
 | | Crystal structure of the Legionella pneumophila type II secretion system substrate NttE | | Descriptor: | 1,2-ETHANEDIOL, NttE | | Authors: | Portlock, T.J, Rehman, S, Garnett, J.A. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Dynamics and Cellular Insight Into Novel Substrates of theLegionella pneumophilaType II Secretion System.
Front Mol Biosci, 7, 2020
|
|
4PY7
 
 | | Crystal Structure of Fab 3.1 | | Descriptor: | antibody 3.1 heavy chain, antibody 3.1 light chain | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
3JYC
 
 | |
3JYS
 
 | |
4KS5
 
 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
5Y41
 
 | | Crystal Structure of LIGAND-BOUND NURR1-LBD | | Descriptor: | (13E,15S)-15-hydroxy-9-oxoprosta-10,13-dien-1-oic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sreekanth, R, Lescar, J, Yoon, H.S. | | Deposit date: | 2017-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Nat.Chem.Biol., 2020
|
|
4KDS
 
 | |
3F3T
 
 | | Kinase domain of cSrc in complex with inhibitor RL38 (Type III) | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-naphthalen-1-ylurea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Klueter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-03-03 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new screening assay for allosteric inhibitors of cSrc
Nat.Chem.Biol., 5, 2009
|
|
6FT2
 
 | |
3AXX
 
 | |
3W81
 
 | | Human alpha-l-iduronidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5HC3
 
 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
3AYC
 
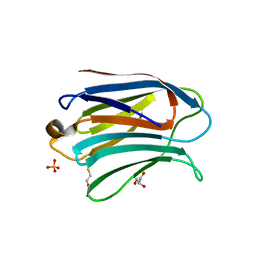 | | Crystal structure of galectin-3 CRD domian complexed with GM1 pentasaccharide | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Galectin-3, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
2XK7
 
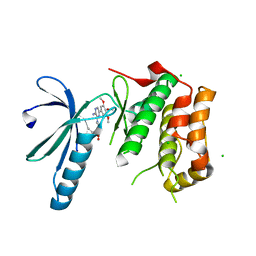 | | Structure of Nek2 bound to aminopyrazine compound 23 | | Descriptor: | (3R,4R)-1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-3-ethylpiperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
3KC3
 
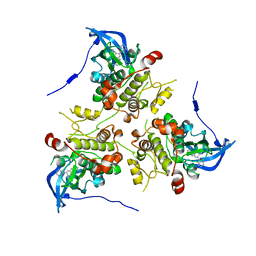 | | MK2 complexed to inhibitor N4-(7-(benzofuran-2-yl)-1H-indazol-5-yl)pyrimidine-2,4-diamine | | Descriptor: | MAP kinase-activated protein kinase 2, N~4~-[7-(1-benzofuran-2-yl)-1H-indazol-5-yl]pyrimidine-2,4-diamine | | Authors: | Argiriadi, M.A, Talanian, R.V, Borhani, D.W. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2,4-Diaminopyrimidine MK2 inhibitors. Part I: Observation of an unexpected inhibitor binding mode.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1P2D
 
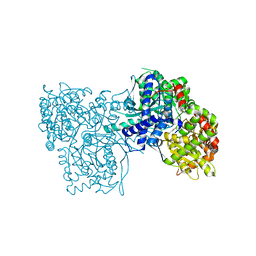 | | Crystal Structure of Glycogen Phosphorylase B in complex with Beta Cyclodextrin | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
5YA2
 
 | | Crystal structure of LsrK-HPr complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
4KVY
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
6MOU
 
 | |
5PAQ
 
 | | Crystal Structure of Factor VIIa in complex with 2-[(1-aminoisoquinolin-6-yl)amino]-2-(3-ethoxy-4-propan-2-yloxyphenyl)-1-(2-phenylpyrrolidin-1-yl)ethanone | | Descriptor: | 2-[(1-aminoisoquinolin-6-yl)amino]-2-(3-ethoxy-4-propan-2-yloxyphenyl)-1-(2-phenylpyrrolidin-1-yl)ethanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
7F3M
 
 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
2XIR
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with PF- 00337210 (N,2-dimethyl-6-(7-(2-morpholinoethoxy)quinolin-4-yloxy) benzofuran-3-carboxamide) | | Descriptor: | N,2-DIMETHYL-6-{[7-(2-MORPHOLIN-4-YLETHOXY)QUINOLIN-4-YL]OXY}-1-BENZOFURAN-3-CARBOXAMIDE, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | Authors: | McTigue, M, Wickersham, J, Pinko, C, Hong, Y, Marrone, T. | | Deposit date: | 2010-06-30 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of the Selective Vegfr Inhibitor Pf- 00337210
To be Published
|
|
4CD8
 
 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4TLR
 
 | | NS5b in complex with lactam-thiophene carboxylic acids | | Descriptor: | 3-{(2R,5R)-5-cyclohexyl-2-[(2R)-2-hydroxypropyl]-3-oxomorpholin-4-yl}-5-(3,3-dimethylbut-1-yn-1-yl)thiophene-2-carboxylic acid, 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, NS5b | | Authors: | Chopra, R. | | Deposit date: | 2014-05-30 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design and synthesis of lactam-thiophene carboxylic acids as potent hepatitis C virus polymerase inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
