5P7U
 
 | |
5P8D
 
 | |
5P8S
 
 | |
5TGA
 
 | |
4MGK
 
 | | Selective activation of Epac1 and Epac2 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
4MGZ
 
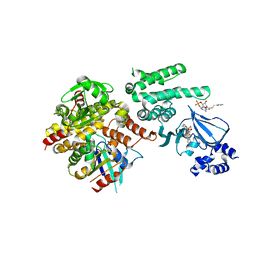 | | Selective activation of Epac1 and Epac2 | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-[6-amino-8-(benzylsulfanyl)-9H-purin-9-yl]-2-sulfanyltetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-7-ol 2-oxide, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
2CSJ
 
 | |
5M0B
 
 | |
7MLX
 
 | |
1Q6X
 
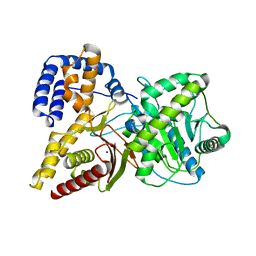 | | Crystal structure of rat choline acetyltransferase | | Descriptor: | SODIUM ION, choline O-acetyltransferase | | Authors: | Cai, Y, Rodgers, D.W. | | Deposit date: | 2003-08-14 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Choline acetyltransferase structure reveals distribution of mutations that cause motor disorders.
Embo J., 23, 2004
|
|
1QXH
 
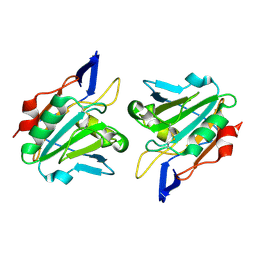 | |
1Q8T
 
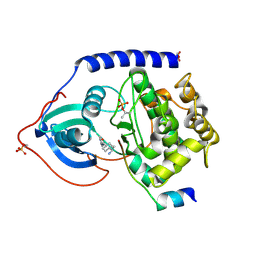 | | The Catalytic Subunit of cAMP-dependent Protein Kinase (PKA) in Complex with Rho-kinase Inhibitor Y-27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
1QD8
 
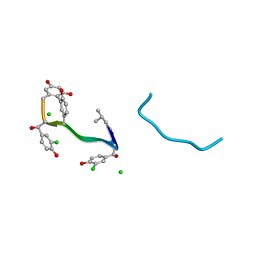 | | COMPLEX OF VANCOMYCIN WITH N-ACETYL GLYCINE | | Descriptor: | ACETYLAMINO-ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-15 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin Binding to Low-Affinity Ligands: Delineating a Minimum Set of Interactions Necessary for High-Affinity Binding.
J.Med.Chem., 42, 1999
|
|
5FPQ
 
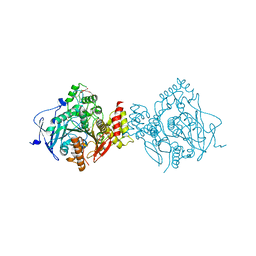 | | Structure of Homo sapiens acetylcholinesterase phosphonylated by sarin. | | Descriptor: | ACETYLCHOLINESTERASE, PENTAETHYLENE GLYCOL | | Authors: | Allgardsson, A, Berg, L, Akfur, C, Hornberg, A, Worek, F, Linusson, A, Ekstrom, F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Prereaction Complex between the Nerve Agent Sarin, its Biological Target Acetylcholinesterase, and the Antidote Hi-6.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2CT3
 
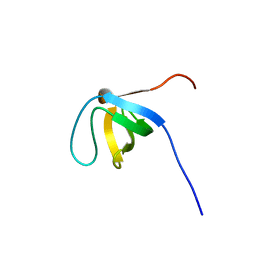 | | Solution Structure of the SH3 domain of the Vinexin protein | | Descriptor: | Vinexin | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the SH3 domain of the Vinexin protein
To be Published
|
|
6B5N
 
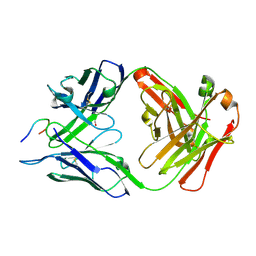 | | Structure of PfCSP peptide 25 with human protective antibody CIS43 | | Descriptor: | CIS43 Fab Heavy chain, CIS43 Fab Light chain, pfCSP peptide 25: ASN-VAL-ASP-PRO-ASN-ALA-ASN-PRO-ASN-VAL-ASP | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6B6W
 
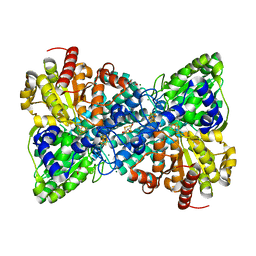 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase, as-isolated (protein batch 2), oxidized C-cluster | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase.
Elife, 7, 2018
|
|
5M71
 
 | |
6B6V
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase, as-isolated (protein batch 1), canonical C-cluster | | Descriptor: | Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase.
Elife, 7, 2018
|
|
6B9X
 
 | | Crystal structure of Ragulator | | Descriptor: | Hepatitis B virus x interacting protein, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | SU, M.-Y, Hurley, J.H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex.
Mol. Cell, 68, 2017
|
|
6B5O
 
 | | Structure of PfCSP peptide 29 with human protective antibody CIS43 | | Descriptor: | CIS43 Fab Heavy chain, CIS43 Fab Light chain, PfCSP peptide 29: ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-ASN | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
2D2Z
 
 | | Crystal structure of Soluble Form Of CLIC4 | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Li, Y.F, Li, D.F, Wang, D.C. | | Deposit date: | 2005-09-21 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric structure of the wild soluble chloride intracellular ion channel CLIC4 observed in crystals
Biochem.Biophys.Res.Commun., 343, 2006
|
|
1QVA
 
 | | YEAST INITIATION FACTOR 4A N-TERMINAL DOMAIN | | Descriptor: | INITIATION FACTOR 4A | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase
RNA, 5, 1999
|
|
6B6Y
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase, dithionite-reduced then oxygen-exposed (protein batch 2), oxidized C-cluster | | Descriptor: | Carbon monoxide dehydrogenase, FE2/S2 (INORGANIC) CLUSTER, Fe(4)-Ni(1)-S(4) cluster, ... | | Authors: | Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase.
Elife, 7, 2018
|
|
5M6Y
 
 | | Cocrystal structure of cAMP-dependent Protein Kinase (PKA) in complex with a methylisoquinoline Fasudil-derivative | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)-4-methyl-isoquinoline, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2016-10-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Cocrystal structure of cAMP-dependent Protein Kinase (PKA) in complex with differently methylated Fasudil-derived ligands
To be Published
|
|
