5VCW
 
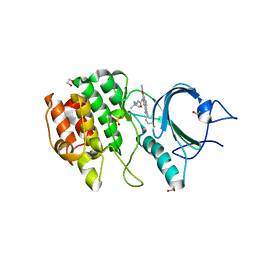 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
7ZWD
 
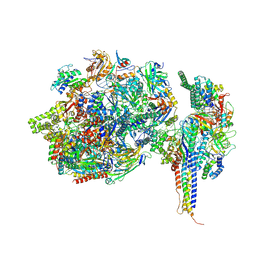 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1LA4
 
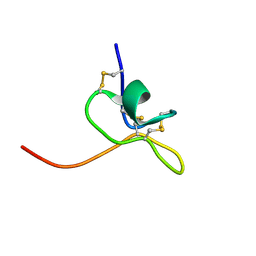 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
8T3A
 
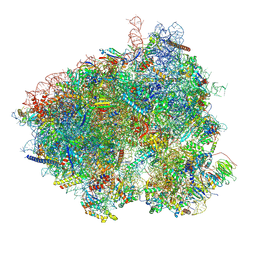 | | Hypomethylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, sordarin, and hibernating factor Los2 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Li, H. | | Deposit date: | 2023-06-07 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Regulation of Translation by Ribosome RNA 2'-O-Methylation
To be Published
|
|
7U50
 
 | |
8T2Z
 
 | | Hypomethylated yeast 80S bound with cycloheximide, P-site tRNA, and A-site tRNA, messenger RNA, POST | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zhao, Y, Li, H. | | Deposit date: | 2023-06-07 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of Hypopseudouridylated Ribosome in IRES-mediated Translation
To Be Published
|
|
8T30
 
 | | Hypomethylated yeast 80S bound with cycloheximide, unmodified U2921, mid rotated | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zhao, Y, Li, H. | | Deposit date: | 2023-06-07 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Regulation of Translation by Ribosome RNA 2'-O-Methylation
To be published
|
|
7GVM
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 4 at 1.40 MGy X-ray dose | | Descriptor: | 5-[(2,5-dichloropyridin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7TTH
 
 | | Human potassium-chloride cotransporter 1 in inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, POTASSIUM ION, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8T3E
 
 | | Hypomethylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure IV | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Li, H. | | Deposit date: | 2023-06-07 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of Hypopseudouridylated Ribosome in IRES-mediated Translation Initiation
To be Published
|
|
6BFX
 
 | | BACE crystal structure with hydroxy pyrrolidine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1S,2S)-3-(3,5-difluorophenyl)-1-[(3R,5S,6R)-6-(2,2-dimethylpropoxy)-5-methylmorpholin-3-yl]-1-hydroxypropan-2-yl}acetamide | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
4O8J
 
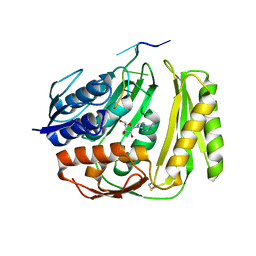 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii, in complex with rACAAA3'phosphate and adenine. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, RNA, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
7GXP
 
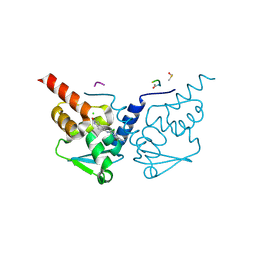 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 9 at 1.42 MGy X-ray dose. | | Descriptor: | (8S)-5-chloro-7-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
6GYO
 
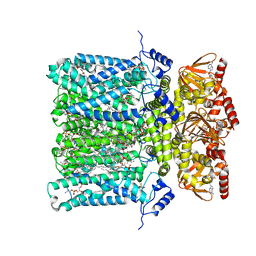 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel in complex with cAMP | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
3F29
 
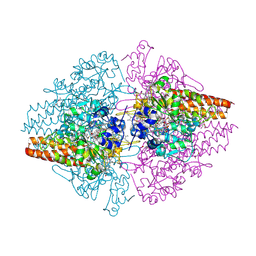 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-10-29 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
3F40
 
 | |
6BFE
 
 | | BACE crystal structure with hydroxy pyrrolidine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(1R,2S)-1-[(2R,4R)-4-(cyclohexylmethoxy)pyrrolidin-2-yl]-3-(3,5-difluorophenyl)-1-hydroxypropan-2-yl]acetamide | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
8JPC
 
 | | cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 2 | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-adrenergic receptor kinase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
6E4A
 
 | | Crystal structure of human BRD4(1) in complex with CN750 | | Descriptor: | 5-(4-{[(3-chlorophenyl)methyl]amino}-2-{4-[2-(dimethylamino)ethyl]piperazin-1-yl}quinazolin-6-yl)-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Fontano, E, White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and lead identification of quinazoline-based BRD4 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7U8D
 
 | | FKBP12 mutant V55G bound to Rapa*-3Z | | Descriptor: | (3S,5Z,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5-(ethoxyimino)-9,27-dihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Wassarman, D.R, Shokat, K.M. | | Deposit date: | 2022-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Tissue-restricted inhibition of mTOR using chemical genetics.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5I2E
 
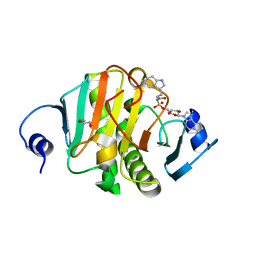 | | Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) with Bound Sulfamate Inhibitor 3a:3-(5-O-{[3-(1H-indol-3-yl)propanoyl]sulfamoyl}-beta-D-ribofuranosyl)-3H-imidazo[2,1-i]purine | | Descriptor: | 3-(5-O-{[3-(1H-indol-3-yl)propanoyl]sulfamoyl}-beta-D-ribofuranosyl)-3H-imidazo[2,1-i]purine, GLYCEROL, Histidine triad nucleotide-binding protein 1 | | Authors: | Strom, A.M, Finzel, B.C, Wagner, C.R. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of Sulfamide and Sulfamate Nucleotidomimetic Inhibitors of hHint1.
Acs Med.Chem.Lett., 7, 2016
|
|
7GV7
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 3 at 1.45 MGy X-ray dose | | Descriptor: | 5-[(5,6-dichloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
3IXL
 
 | |
6TCA
 
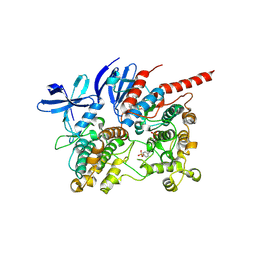 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
3F7S
 
 | |
