6UUM
 
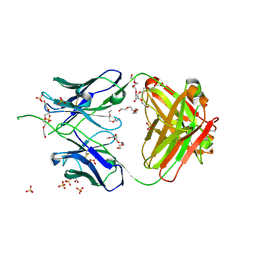 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
7HPD
 
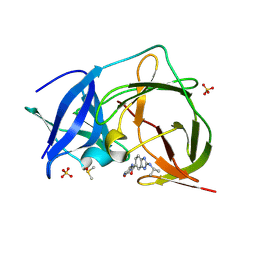 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0015216-001 | | Descriptor: | DIMETHYL SULFOXIDE, N-(5-carbamoyl-1H-pyrrol-3-yl)-2-[(propan-2-yl)amino]-1H-1,3-benzimidazole-6-carboxamide, SULFATE ION, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Lee, A, Kenton, N, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8CHF
 
 | | cryo-EM Structure of Craf:14-3-3:Mek1 | | Descriptor: | 14-3-3 protein zeta isoform X1, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Dedden, D, Ulrich, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
1WAD
 
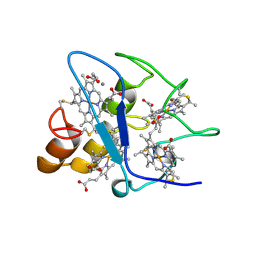 | | CYTOCHROME C3 WITH 4 HEME GROUPS AND ONE CALCIUM ION | | Descriptor: | CALCIUM ION, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Morais, J, Coelho, R, Carrondo, M.A, Wilson, K, Dauter, Z, Sieker, L. | | Deposit date: | 1996-01-10 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytochrome c3 from Desulfovibrio gigas: crystal structure at 1.8 A resolution and evidence for a specific calcium-binding site.
Protein Sci., 5, 1996
|
|
6BRY
 
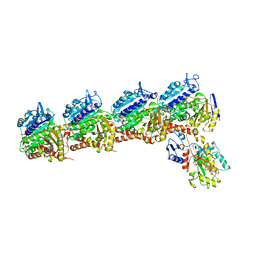 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
9HSN
 
 | |
7U9Y
 
 | |
5WRD
 
 | | Crystal structure of LC3B in complex with FYCO1 LIR | | Descriptor: | GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FYVE and coiled-coil domain-containing protein 1 | | Authors: | Sakurai, S, Ohto, U, Shimizu, T. | | Deposit date: | 2016-12-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of mouse LC3B in complex with the FYCO1 LIR reveals the importance of the flanking region of the LIR motif
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5TXT
 
 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
4GKR
 
 | |
9HSV
 
 | |
7BIY
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-175 | | Descriptor: | 1-(4-methanoylphenyl)carbonylpiperidine-4-carbonitrile, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
5C2Y
 
 | | Crystal structure of the Saccharomyces cerevisiae Rtr1 (regulator of transcription) | | Descriptor: | GLYCEROL, RNA polymerase II subunit B1 CTD phosphatase RTR1, SULFATE ION, ... | | Authors: | Yogesha, S.D, Irani, S, Zhang, Y.J. | | Deposit date: | 2015-06-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase.
Sci.Signal., 9, 2016
|
|
3G7R
 
 | | Crystal structure of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-10 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and ligand specificity of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
9HSP
 
 | |
7F3M
 
 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
1BEA
 
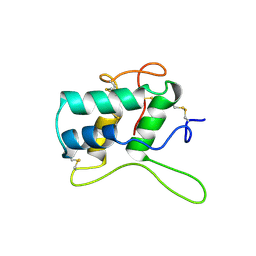 | | BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
9HSD
 
 | |
3K8Q
 
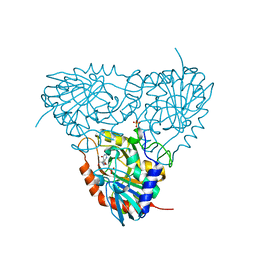 | | Crystal structure of human purine nucleoside phosphorylase in complex with SerMe-Immucillin H | | Descriptor: | 7-({[2-hydroxy-1-(hydroxymethyl)ethyl]amino}methyl)-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ho, M, Almo, S.C, Scharmm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human nucleoside phosphorylase in complex with SerMe-ImmH
Proc.Natl.Acad.Sci.USA, 2010
|
|
5LCJ
 
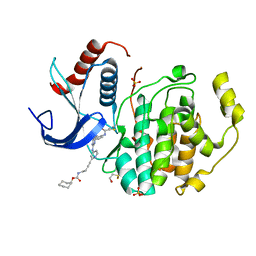 | | In-Gel Activity-Based Protein Profiling of a Clickable Covalent Erk 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[5-chloranyl-4-[[2-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
1BEY
 
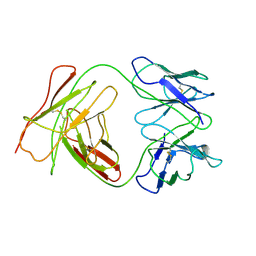 | |
9F4N
 
 | | UP1 in complex with Z137811222 | | Descriptor: | 1-[4-(3-phenylpropyl)piperazin-1-yl]ethan-1-one, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNPA1 via cryptic pockets mapping coupled with X-ray fragment screening.
J.Biol.Chem., 301, 2025
|
|
5U7J
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 5-[2-(2-methoxyphenyl)ethoxy]-3-(2-methylpropyl)[1,2,4]triazolo[4,3-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
4UQC
 
 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum at 1.30 A resolution | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-06-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5ICE
 
 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and norlaudanosoline | | Descriptor: | (1S)-1-(3,4-dihydroxybenzyl)-1,2,3,4-tetrahydroisoquinoline-6,7-diol, (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
