9CZU
 
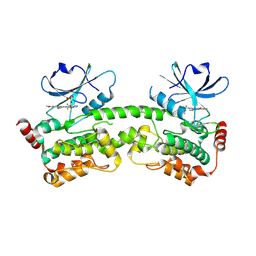 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 9 | | Descriptor: | 6-methoxy-4-[(methylamino)methyl]-2-(6-{4-[(2S)-4,4,4-trifluorobutan-2-yl]-4H-1,2,4-triazol-3-yl}pyridin-2-yl)-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9CZT
 
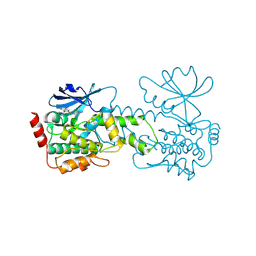 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 6 | | Descriptor: | 4-(aminomethyl)-2-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9CZW
 
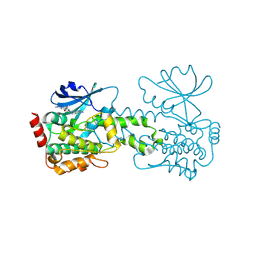 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 13 | | Descriptor: | 6-(dimethylamino)-4-[(methylamino)methyl]-2-[6-(4-propyl-4H-1,2,4-triazol-3-yl)pyridin-2-yl]-2,3-dihydro-1H-pyrrolo[3,4-c]pyridin-1-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, Mc Tigue, M. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9D00
 
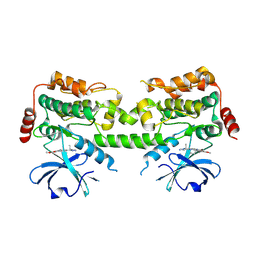 | | HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 53 | | Descriptor: | 4-[(1R)-1-aminopropyl]-6-methoxy-2-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2024-08-05 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Discovery of PF-07265028, A Selective Small Molecule Inhibitor of Hematopoietic Progenitor Kinase 1 (HPK1) for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
5ZEE
 
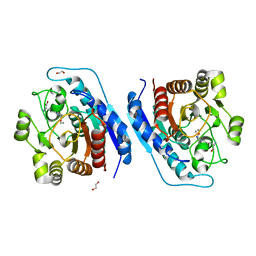 | | Crystal structure of Entamoeba histolytica Arginase in complex with N(omega)-hydroxy-L-arginine (NOHA) at 1.74 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, MANGANESE (II) ION, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
6F3U
 
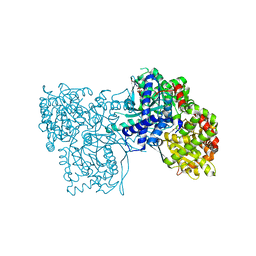 | | The crystal structure of Glycogen Phosphorylase in complex with 10h | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(5-naphthalen-1-yl-4~{H}-1,2,4-triazol-3-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Gkerdi, A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
5N5I
 
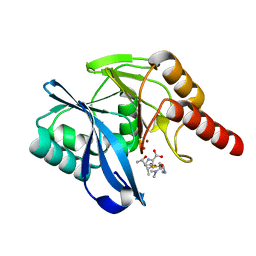 | | Crystal Structure of VIM-1 metallo-beta-lactamase in complex with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
5DWF
 
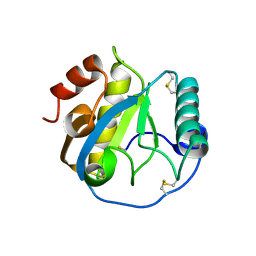 | | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Singh, P.K, Yadav, S.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution
To Be Published
|
|
4ZYF
 
 | |
5E1M
 
 | | Crystal structure of NTMT1 in complex with PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
3T9T
 
 | | Crystal structure of BTK mutant (F435T,K596R) complexed with Imidazo[1,5-a]quinoxaline | | Descriptor: | (2Z)-4-(dimethylamino)-N-{7-fluoro-4-[(2-methylphenyl)amino]imidazo[1,5-a]quinoxalin-8-yl}-N-methylbut-2-enamide, GLYCEROL, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5E2A
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
3VBT
 
 | |
3F5E
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 2'F-3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
3KZT
 
 | |
6E8X
 
 | | CA IX mimic Complexed with Steroidal Sulfamate Compound STX 140 | | Descriptor: | (13alpha,17alpha)-2-methoxyestra-1,3,5,7,9,11-hexaene-3,17-diyl disulfamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, Mckenna, R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 3,17 beta-Bis-sulfamoyloxy-2-methoxyestra-1,3,5(10)-triene and Nonsteroidal Sulfamate Derivatives Inhibit Carbonic Anhydrase IX: Structure-Activity Optimization for Isoform Selectivity.
J. Med. Chem., 62, 2019
|
|
6QYK
 
 | | Structure of MBP-Mcl-1 in complex with compound 7a | | Descriptor: | (2~{R})-2-[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]oxypropanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8XXA
 
 | |
4YWQ
 
 | | Crystal structure of the ROQ domain of human Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ROQ domain of human Roquin-1
To be Published
|
|
7XJV
 
 | | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form | | Descriptor: | Alpha-1,3-mannosyltransferase MNT2, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Hira, D, Kadooka, C, Oka, T. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form
To Be Published
|
|
7AU5
 
 | | Tubulin-noscapine-analogue-14e complex | | Descriptor: | (5~{R})-5-[(1~{S})-4,5-dimethoxy-1,3-dihydro-2-benzofuran-1-yl]-~{N}-ethyl-4-methoxy-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinoline-6-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yong, C, Devine, S.M, Abel, A.-C, Muthiah, D, Gao, X, Callaghan, R, Capuano, B, Steinmetz, M.O, Prota, A.E, Scammels, P.J. | | Deposit date: | 2020-11-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1,3-Benzodioxole-Modified Noscapine Analogues: Synthesis, Antiproliferative Activity, and Tubulin-Bound Structure.
Chemmedchem, 16, 2021
|
|
3T3C
 
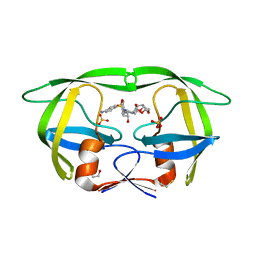 | | Structure of HIV PR resistant patient derived mutant (comprising 22 mutations) in complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, HIV-1 protease, ... | | Authors: | Rezacova, P, Kozisek, M, Konvalinka, J, Saskova, K.G. | | Deposit date: | 2011-07-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in HIV-1 gag and pol Compensate for the Loss of Viral Fitness Caused by a Highly Mutated Protease.
Antimicrob.Agents Chemother., 56, 2012
|
|
9CMY
 
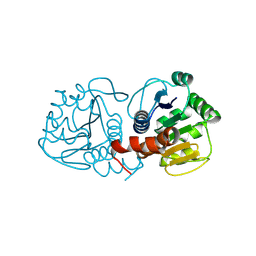 | | Human DJ-1, 6.5-18.5 min mixing with methylglyoxal, fixed target serial crystallography | | Descriptor: | Protein deglycase DJ-1 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-07-15 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
3F5A
 
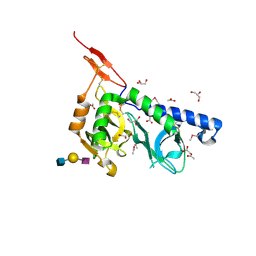 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
6QYO
 
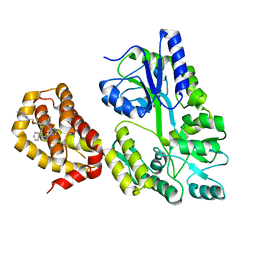 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
