4ZDR
 
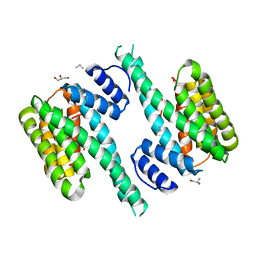 | | Crystal structure of 14-3-3[zeta]-LKB1 fusion protein | | Descriptor: | 14-3-3 protein zeta/delta,GGSGGS linker,Serine/threonine-protein kinase STK11, GLYCEROL, PROPANE, ... | | Authors: | Ding, S, Shi, Z.B. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structure of the 14-3-3 zeta-LKB1 fusion protein provides insight into a novel ligand-binding mode of 14-3-3.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6KEY
 
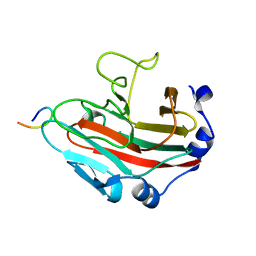 | |
8GCK
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | Descriptor: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36823535 Å) | | Cite: | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
6I7C
 
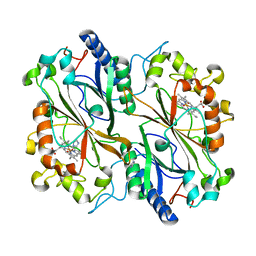 | | Dye type peroxidase Aa from Streptomyces lividans: imidazole complex | | Descriptor: | Deferrochelatase/peroxidase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno-Chicano, T, Ebrahim, A.E, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6I7F
 
 | | Dehaloperoxidase B from Amphitrite ornata - complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moreno-Chicano, T, Ebrahim, A.E, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
7PNE
 
 | |
7PNG
 
 | |
4XOU
 
 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
5L8I
 
 | | crystal structure of human FABP6 apo-protein | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, Gastrotropin, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
5L8N
 
 | | crystal structure of human FABP6 protein with fragment 1 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 5,6-dimethyl-1~{H}-benzimidazol-2-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
6XCL
 
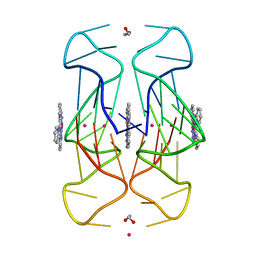 | | Crystal Structure of human telomeric DNA G-quadruplex in complex with a novel platinum(II) complex. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), POTASSIUM ION, ... | | Authors: | Miron, C.E, van Staalduinen, L.M, Jia, Z, Petitjean, A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Going Platinum to the Tune of a Remarkable Guanine Quadruplex Binder: Solution- and Solid-State Investigations.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4D0F
 
 | | Human Notch1 EGF domains 11-13 mutant T466A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6XVC
 
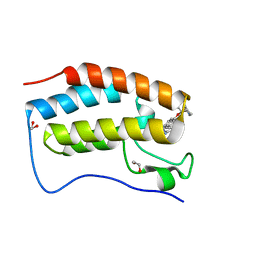 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 1 | | Descriptor: | (4~{R})-4-[(1~{R})-1-[7-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)quinolin-5-yl]oxyethyl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8HB1
 
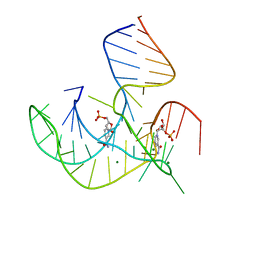 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
4CUE
 
 | | Human Notch1 EGF domains 11-13 mutant T466V | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HBA
 
 | |
8HB8
 
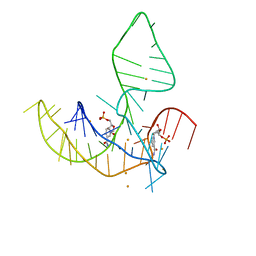 | |
8HB3
 
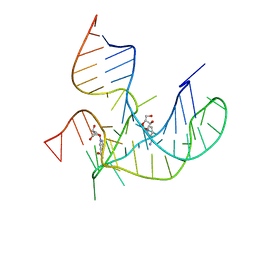 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
1A5M
 
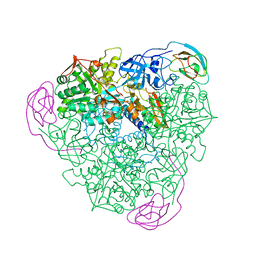 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5O
 
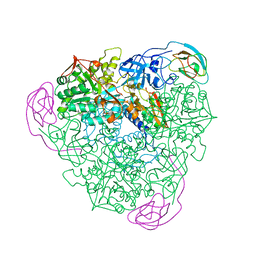 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5N
 
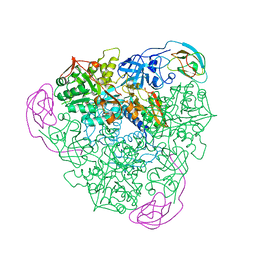 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5K
 
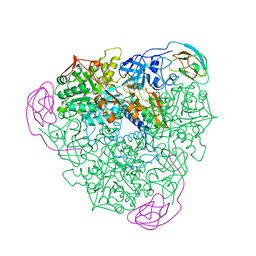 | | K217E VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5L
 
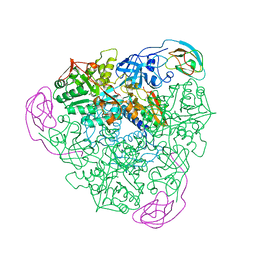 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
4D0E
 
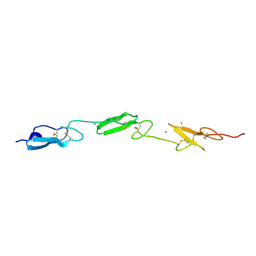 | | Human Notch1 EGF domains 11-13 mutant GlcNAc-fucose disaccharide modified at T466 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CUD
 
 | | Human Notch1 EGF domains 11-13 mutant fucosylated at T466 | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, alpha-L-fucopyranose | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
