5OU1
 
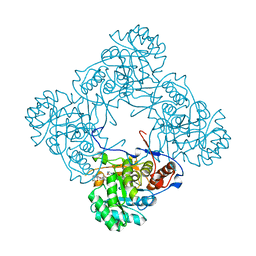 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 (7759844) | | Descriptor: | (2~{S})-~{N}-(4-iodophenyl)-2-(4-methoxyphenoxy)propanamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
5OU3
 
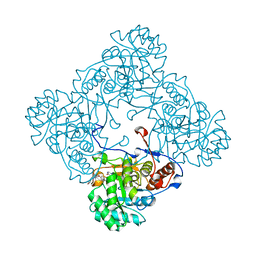 | | M. thermoresistible IMPDH in complex with IMP and Compound 31 (AT080) | | Descriptor: | (2~{S})-~{N}-[5-(4-bromophenyl)-1~{H}-imidazol-2-yl]-2-[4-(1-methylimidazol-4-yl)phenoxy]propanamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
6D4V
 
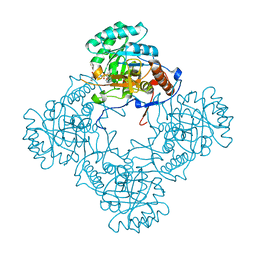 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 22 (VCC061422) | | Descriptor: | 2-cyclohexyl-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4R
 
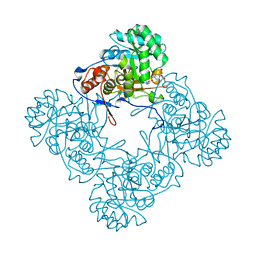 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 18 (VCC399134) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, hydroxy(3-{4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carbonyl}phenyl)oxoammonium | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4U
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 27 (VCC663664) | | Descriptor: | 2-(2,4-dimethoxyphenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
4IX2
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP.
To be Published
|
|
4FO4
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP and mycophenolic acid | | Descriptor: | INOSINIC ACID, Inosine 5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP and mycophenolic acid.
To be Published
|
|
7OJ2
 
 | | Bacillus subtilis IMPDH in complex with Ap4A | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Giammarinaro, P.I, Bange, G. | | Deposit date: | 2021-05-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Nat Microbiol, 7, 2022
|
|
6D4T
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 45 (VCC117054) | | Descriptor: | (2S)-N-(2H-1,3-benzodioxol-5-yl)-4-[(isoquinolin-5-yl)sulfonyl]-2-methylpiperazine-1-carboxamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4S
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 37 (VCC670597) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, N-(2,3-dichlorophenyl)-4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carboxamide | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | Descriptor: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
2A1Y
 
 | | Crystal Structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A Resolution. | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A resolution.
To be Published
|
|
6D4W
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 35 (VCC620637) | | Descriptor: | 2-(4-fluorophenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4Q
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 14 (VCC900455) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, cycloheptyl{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}methanone | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
2BLE
 
 | | Structure of human guanosine monophosphate reductase GMPR1 in complex with GMP | | Descriptor: | GMP REDUCTASE I, GUANOSINE-5'-MONOPHOSPHATE, SODIUM ION | | Authors: | Bunkoczi, G, Haroniti, A, Ng, S, von Delft, F, Oppermann, U, Arrowsmith, C, Sundstrom, M, Edwards, A, Gileadi, O. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Guanosine Monophosphate Reductase Gmpr1 in Complex with Gmp
To be Published
|
|
4Q33
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110 | | Descriptor: | 4-[(1R)-1-[1-(4-chlorophenyl)-1,2,3-triazol-4-yl]ethoxy]-1-oxidanyl-quinoline, ACETIC ACID, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110
TO BE PUBLISHED
|
|
4MJM
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2544 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames
To be Published, 2013
|
|
4QM1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67 | | Descriptor: | 2-(3-methyl-4-oxo-3,4-dihydrophthalazin-1-yl)-N-(6,7,8,9-tetrahydrodibenzo[b,d]furan-2-yl)acetamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Mandapati, K, Gollapalli, D, Gorla, S.K, Zhang, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7964 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67
To be Published, 2014
|
|
4MYX
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-5-{[(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)carbamoyl]amino}benzamide, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32
To be Published
|
|
4MYA
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110 | | Descriptor: | 4-{(1R)-1-[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]ethoxy}quinolin-2(1H)-one, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110
To be Published
|
|
4R7J
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1172 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni
To be Published, 2014
|
|
4QQ3
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP | | Descriptor: | CHLORIDE ION, Inosine-5'-monophosphate dehydrogenase, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from vibrio cholerae, deletion mutant, in complex with xmp
To be Published
|
|
4MY1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68 | | Descriptor: | 1-(4-bromophenyl)-3-(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)urea, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68
To be Published
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5893 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
