8QV9
 
 | | Hexameric HIV-1 CA in complex with DDD01829021 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromanyl-3-(phenylmethyl)-1~{H}-benzimidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV4
 
 | | Hexameric HIV-1 CA in complex with DDD01728503 | | Descriptor: | 1,2-ETHANEDIOL, Spacer peptide 1, ethyl 2-(3-oxidanylidene-2,4-dihydroquinoxalin-1-yl)ethanoate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV1
 
 | | Hexameric HIV-1 CA in complex with DDD01728505 | | Descriptor: | Spacer peptide 1, methyl 2-(2-oxidanylidene-1~{H}-quinolin-4-yl)ethanoate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUY
 
 | | Hexameric HIV-1 CA in complex with DDD01728501 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylphenyl)methyl]-1~{H}-quinoxaline-2,3-dione, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUX
 
 | | Hexameric HIV-1 CA in complex with DDD00100333 | | Descriptor: | 1,2-ETHANEDIOL, 4-benzyl-3,4-dihydroquinoxalin-2(1H)-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUW
 
 | | Hexameric HIV-1 CA in complex with DDD01044153 | | Descriptor: | (4~{R})-7-oxidanyl-4-phenyl-3,4-dihydro-1~{H}-quinolin-2-one, 1,2-ETHANEDIOL, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUL
 
 | | Hexameric HIV-1 CA in complex with DDD00100555 | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUK
 
 | | Hexameric HIV-1 CA in complex with DDD00100439 | | Descriptor: | (phenylmethyl) 3-oxidanylidenepiperazine-1-carboxylate, 1,2-ETHANEDIOL, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUJ
 
 | | Hexameric HIV-1 CA in complex with DDD00100452 | | Descriptor: | 1,2-ETHANEDIOL, 3-(phenylmethyl)-1~{H}-imidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUI
 
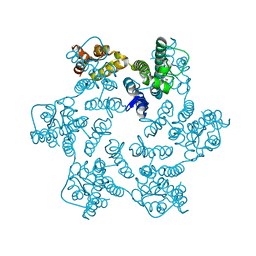 | | Hexameric HIV-1 CA in complex with DDD00024969 | | Descriptor: | Spacer peptide 1, ethyl (3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)acetate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUH
 
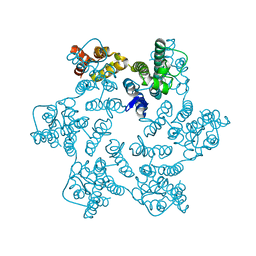 | | Hexameric HIV-1 CA in complex with DDD00057456 | | Descriptor: | 4-methylquinolin-2-ol, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUB
 
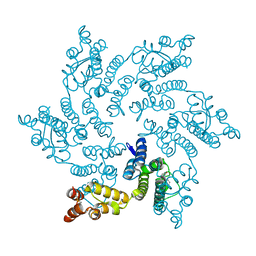 | | Hexameric HIV-1 CA in complex with DDD00074110 | | Descriptor: | (1~{S})-1-phenyl-2,4-dihydro-1~{H}-isoquinolin-3-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8PUH
 
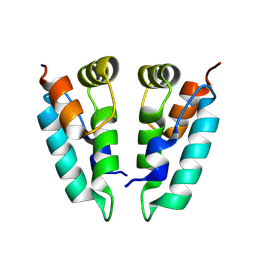 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PT5
 
 | | ERK2 covelently bound to RU187 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PT3
 
 | | ERK2 covelently bound to RU77 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PT1
 
 | | ERK2 covelently bound to RU76 cyclohexenone based inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ~{O}1-methyl ~{O}3-(phenylmethyl) (1~{S},3~{R})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PT0
 
 | | ERK2 covelently bound to RU75 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PSY
 
 | | ERK2 covelently bound to RU68 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{S},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PSW
 
 | | ERK2 covalently bound to RU67 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PST
 
 | | ERK2 covelently bound to RU60 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{S})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PSR
 
 | | ERK2 covalently bound to SynthRevD-12-opt artificial peptide | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Gogl, G, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8OTF
 
 | |
8GDV
 
 | |
8G6O
 
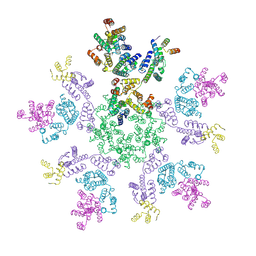 | | HIV-1 capsid lattice bound to IP6 and Lenacapavir | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
