6QXJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 6a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]amino]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
5D03
 
 | |
4Z77
 
 | |
7UGP
 
 | |
7MMZ
 
 | |
3VBX
 
 | |
3FBP
 
 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
6PNN
 
 | |
9EN2
 
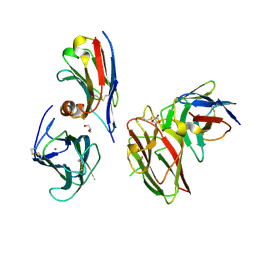 | | Crystal structure of the metalloproteinase enhancer PCPE-1 complexed with nanobodies VHH-H4 and VHH-I5 | | Descriptor: | CALCIUM ION, GLYCEROL, Procollagen C-endopeptidase enhancer 1, ... | | Authors: | Lagoutte, P, Gueguen-Chaignon, V, Bourhis, J.-M, Vadon-Le Goff, S. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mono- and Bi-specific Nanobodies Targeting the CUB Domains of PCPE-1 Reduce the Proteolytic Processing of Fibrillar Procollagens.
J.Mol.Biol., 436, 2024
|
|
4QRL
 
 | |
6TJ8
 
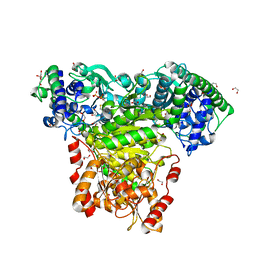 | | Escherichia coli transketolase in complex with cofactor analog 2'-methoxythiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-azanyl-2-methoxy-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, ... | | Authors: | Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2019-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.921 Å) | | Cite: | Structural basis for antibiotic action of the B 1 antivitamin 2'-methoxy-thiamine.
Nat.Chem.Biol., 16, 2020
|
|
5C16
 
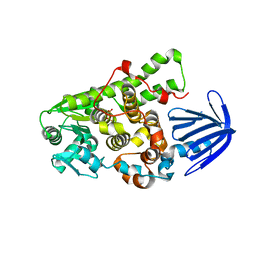 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
7UHF
 
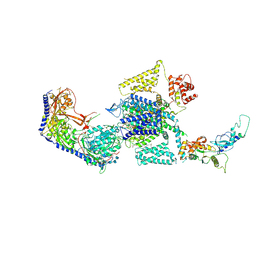 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of cinnarizine at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-(diphenylmethyl)-4-[(2E)-3-phenylprop-2-en-1-yl]piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-03-26 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for pore blockade of human voltage-gated calcium channel Ca v 1.3 by motion sickness drug cinnarizine.
Cell Res., 32, 2022
|
|
6F3J
 
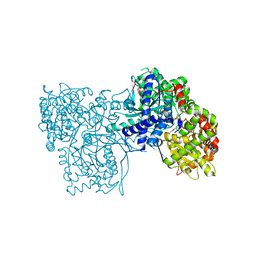 | | The crystal structure of Glycogen Phosphorylase in complex with 10a | | Descriptor: | 4-[4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]phenyl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
3T4E
 
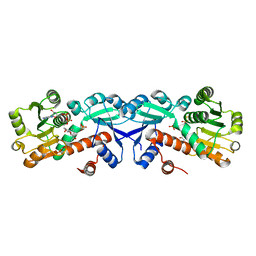 | | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Quinate/shikimate dehydrogenase | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD.
TO BE PUBLISHED
|
|
9BW9
 
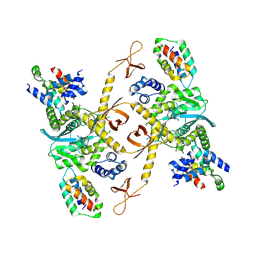 | | Tetrameric Complex of full-length HIV-1 integrase protein bound to the integrase binding domain of LEDGF/p75 | | Descriptor: | Integrase, PC4 and SFRS1-interacting protein | | Authors: | Jing, T, Shan, Z, Lyumkis, D, Biswas, A. | | Deposit date: | 2024-05-21 | | Release date: | 2025-06-25 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Oligomeric HIV-1 integrase structures reveal functional plasticity for intasome assembly and RNA binding.
Nat Commun, 16, 2025
|
|
6F3R
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10c | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[5-(9~{H}-fluoren-2-yl)-4~{H}-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6PNM
 
 | |
5LRM
 
 | | Structure of di-zinc MCR-1 in P41212 space group | | Descriptor: | GLYCEROL, ZINC ION, phosphatidylethanolamine transferase Mcr-1 | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
6F3S
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10d | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-phenylphenyl)-4~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
8VC0
 
 | | HIV-1 CA crosslinked pentamer in complex with GS-CA1 | | Descriptor: | Capsid protein p24, N-[(1S)-1-{(3M)-3-{4-chloro-3-[(cyclopropanesulfonyl)amino]-1-(2,2-difluoroethyl)-1H-indazol-7-yl}-6-[3-(methanesulfonyl)-3-methylbut-1-yn-1-yl]pyridin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Piacentini, J, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2023-12-13 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of disulfide-stabilized HIV-1 CA pentamer in complex with the ultra-potent capsid inhibitor, GS-CA1
To Be Published
|
|
8R74
 
 | | Galectin-1 in complex with thiogalactoside derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-(2-oxidanyl-1,3-thiazol-4-yl)-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-1 | | Authors: | Hakansson, M, Diehl, C, Peterson, K, Zetterberg, F, Nilsson, U. | | Deposit date: | 2023-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of the Selective and Orally Available Galectin-1 Inhibitor GB1908 as a Potential Treatment for Lung Cancer.
J.Med.Chem., 67, 2024
|
|
5LR6
 
 | | Crystal Structure of COMT in complex with [3-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-5-yl]-(4-phenylpiperazin-1-yl)methanone | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of COMT in complex with [3-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-5-yl]-(4-phenylpiperazin-1-yl)methanone
To be published
|
|
4YC2
 
 | |
4YDK
 
 | | Crystal structure of broadly and potently neutralizing antibody C38-VRC16.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
