5CW7
 
 | |
5CZE
 
 | |
5CZF
 
 | |
8XQB
 
 | | Mature virion portal vertex of bacteriophage lambda | | Descriptor: | Capsid decoration protein, Head completion protein, Head-tail connector protein FII, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
7PFP
 
 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7QCG
 
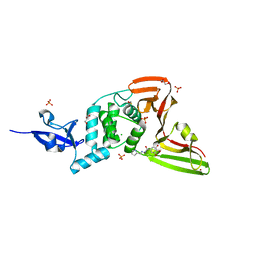 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6MAR
 
 | |
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCK
 
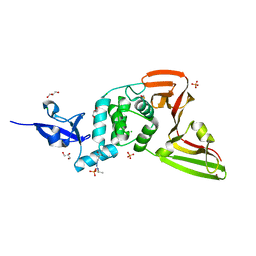 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
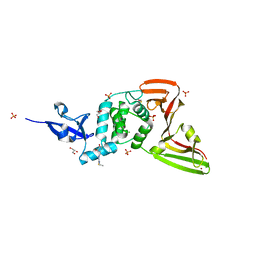 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
3FN3
 
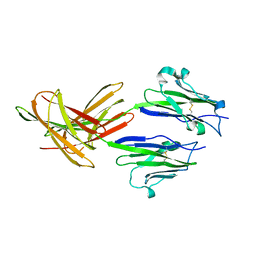 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
8PN9
 
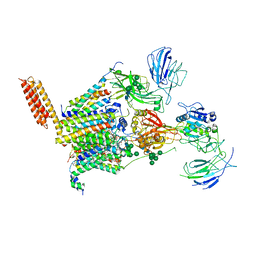 | | Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2023-06-30 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Positive selection CRISPR screens reveal a druggable pocket in an oligosaccharyltransferase required for inflammatory signaling to NF-kappa B.
Cell, 187, 2024
|
|
8R3V
 
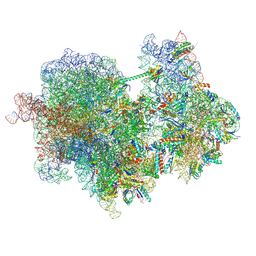 | |
8OEP
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the HPV18 E6 oncoprotein C-terminal peptide | | Descriptor: | Protein E6, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | Deposit date: | 2023-03-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
8SAS
 
 | | CryoEM structure of DH270.5-CH848.10.17 | | Descriptor: | CH848.10.17 gp120, CH848.10.17 gp41, DH270.5 variable heavy chian, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
3HLA
 
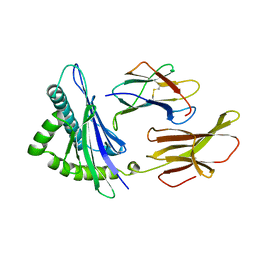 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN A2.1 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2.1) (ALPHA CHAIN) | | Authors: | Saper, M.A, Bjorkman, P.J, Wiley, D.C. | | Deposit date: | 1989-10-06 | | Release date: | 1990-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution.
J.Mol.Biol., 219, 1991
|
|
3O3F
 
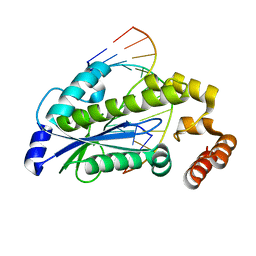 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and magnesium ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
3O3H
 
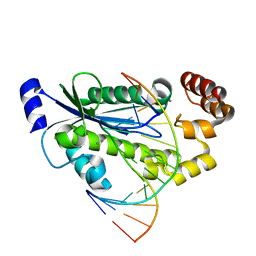 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and manganese ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
6OZC
 
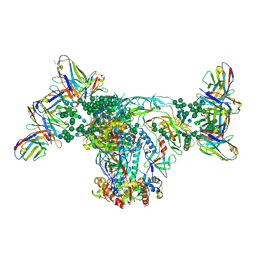 | | BG505 SOSIP.664 with 2G12 Fab2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 Fab Heavy chain, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Structure, 28, 2020
|
|
3J3Y
 
 | |
8TUX
 
 | |
3J3Q
 
 | |
