6GVJ
 
 | | Human Mps1 kinase domain with ordered activation loop | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, GLYCEROL | | Authors: | Roorda, J.C, Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-06-21 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A crystal structure of the human protein kinase Mps1 reveals an ordered conformation of the activation loop.
Proteins, 87, 2019
|
|
7BJ9
 
 | | Structure of Sfh-I with 2-Mercaptomethyl-thiazolidine L-anti-1a | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase, GLYCEROL, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21000588 Å) | | Cite: | 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode.
Acs Infect Dis., 7, 2021
|
|
7BJ8
 
 | | Structure of L1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode.
Acs Infect Dis., 7, 2021
|
|
6I2Y
 
 | | Human STK10 bound to Foretinib | | Descriptor: | N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide, Serine/threonine-protein kinase 10 | | Authors: | Sorrell, F.J, Berger, B.-T, Oerum, S, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Elkins, J.M. | | Deposit date: | 2018-11-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Human STK10 bound to GW683134
To Be Published
|
|
9GXZ
 
 | |
6IC3
 
 | | AL amyloid fibril from a lambda 1 light chain | | Descriptor: | lambda 1 light chain fragment, residues 3-118 | | Authors: | Fritz, G, Faendrich, M, Schmidt, M, Radamaker, L. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis.
Nat Commun, 10, 2019
|
|
5NC0
 
 | | The 0.91 A resolution structure of the L16G mutant of cytochrome c prime from Alcaligenes xylosoxidans, complexed with nitric oxide | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c', HEME C, ... | | Authors: | Strange, R, Hough, M, Antonyuk, S, Rustage, N. | | Deposit date: | 2017-03-02 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Distinguishing Nitro vs Nitrito Coordination in Cytochrome c' Using Vibrational Spectroscopy and Density Functional Theory.
Inorg Chem, 56, 2017
|
|
6I25
 
 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, Aureochrome1-like protein, CHLORIDE ION, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
7BKE
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodisulfide reductase core and mobile arm in conformational state 2, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
8DJV
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
5NC9
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2,6-diamino-N-hydroxyhexanamide | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-oxidanyl-hexanamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
6RCY
 
 | |
6RDC
 
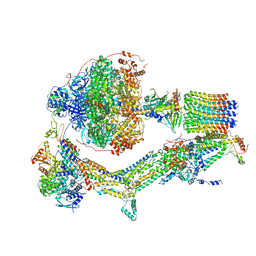 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 2, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7XMH
 
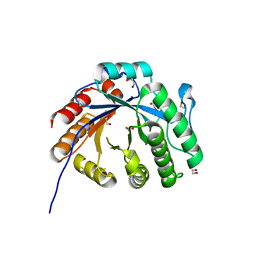 | | Crystal structure of a rice class IIIb chitinase, Oschib2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative class III chitinase | | Authors: | Jun, T, Tomoya, T, Tomoyuki, N, Takayuki, O. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins.
Plant Sci., 326, 2023
|
|
9DPF
 
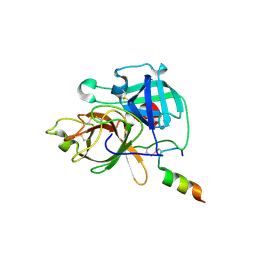 | | Crystal structure of TMPRSS11D S368A complexed with its own zymogen activation motif | | Descriptor: | 1,2-ETHANEDIOL, Transmembrane protease serine 11D, Transmembrane protease serine 11D non-catalytic chain | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Seitova, A, Li, Y, Edwards, A, Benard, F, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-21 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of TMPRSS11D specificity and autocleavage activation.
Nat Commun, 16, 2025
|
|
6RDY
 
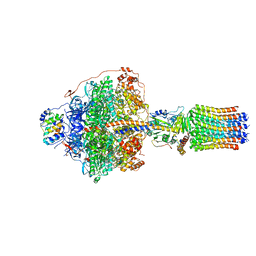 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1F, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
8DBR
 
 | | E. coli ATP synthase imaged in 10mM MgATP State2 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
5ZCB
 
 | | Crystal structure of Alpha-glucosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
6RED
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
8DBT
 
 | | E. coli ATP synthase imaged in 10mM MgATP State2 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
6E0A
 
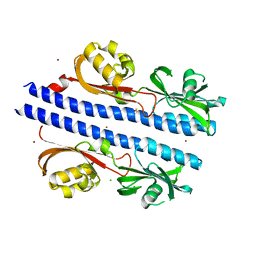 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-07-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
5NND
 
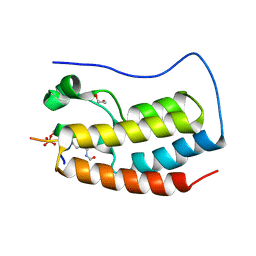 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H3K9ac/K14ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Histone H3 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5HZ8
 
 | | FABP4_3 in complex with 6,8-Dichloro-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid | | Descriptor: | 6,8-dichloro-4-phenyl-2-(piperidin-1-yl)quinoline-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Rudolph, M.G. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Design and synthesis of selective, dual fatty acid binding protein 4 and 5 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
8DBP
 
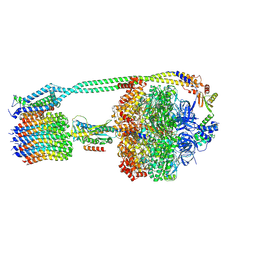 | | E. coli ATP synthase imaged in 10mM MgATP State1 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
6QUP
 
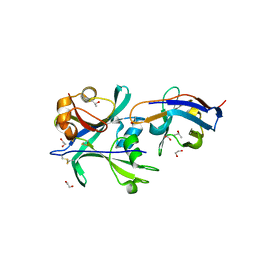 | | Structural signatures in EPR3 define a unique class of plant carbohydrate receptors | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, ... | | Authors: | Wong, J.E, Gysel, K, Birkefeldt, T.G, Vinther, M, Muszynski, A, Azadi, P, Laursen, N.S, Sullivan, J.T, Ronson, C.W, Stougaard, J, Andersen, K.R. | | Deposit date: | 2019-02-28 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Structural signatures in EPR3 define a unique class of plant carbohydrate receptors.
Nat Commun, 11, 2020
|
|
