1PWG
 
 | | Covalent Penicilloyl Acyl Enzyme Complex Of The Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(6S)-6-carboxy-6-(glycylamino)hexanoyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.074 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PWC
 
 | | penicilloyl acyl enzyme complex of the Streptomyces R61 DD-peptidase with penicillin G | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, OPEN FORM - PENICILLIN G | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1SCW
 
 | | TOWARD BETTER ANTIBIOTICS: CRYSTAL STRUCTURE OF R61 DD-PEPTIDASE INHIBITED BY A NOVEL MONOCYCLIC PHOSPHATE INHIBITOR | | Descriptor: | (2Z)-3-{[OXIDO(OXO)PHOSPHINO]OXY}-2-PHENYLACRYLATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Toward Better Antibiotics: Crystallographic Studies of a Novel Class of DD-Peptidase/beta-Lactamase Inhibitors.
Biochemistry, 43, 2004
|
|
1SDE
 
 | | Toward Better Antibiotics: Crystal Structure Of D-Ala-D-Ala Peptidase inhibited by a novel bicyclic phosphate inhibitor | | Descriptor: | 2-[(DIOXIDOPHOSPHINO)OXY]BENZOATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-13 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward better antibiotics: crystallographic studies of a novel class of DD-peptidase/beta-lactamase inhibitors
Biochemistry, 43, 2004
|
|
1PW1
 
 | | Non-Covalent Complex Of Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2S,5R,6R)-6-{[(6R)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, FORMYL GROUP, ... | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PWD
 
 | | Covalent acyl enzyme complex of the Streptomyces R61 DD-peptidase with cephalosporin C | | Descriptor: | (2R)-5-(acetyloxymethyl)-2-[(1R)-1-[[(5R)-5-azanyl-6-oxidanyl-6-oxidanylidene-hexanoyl]amino]-2-oxidanylidene-ethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase precursor | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
6H5O
 
 | |
2XDM
 
 | | Crystal structure of a complex between Actinomadura R39 DD peptidase and a peptidoglycan mimetic boronate inhibitor | | Descriptor: | (D-ALPHA-AMINOPIMELYLAMINO)-D-1-ETHYLBORONIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, ... | | Authors: | Rocaboy, M, Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2010-05-04 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Complex between the Actinomadura R39 Dd-Peptidase and a Peptidoglycan- Mimetic Boronate Inhibitor: Interpretation of a Transition State Analogue in Terms of Catalytic Mechanism.
Biochemistry, 49, 2010
|
|
3BRM
 
 | | Crystal structure of the covalent complex between the Bacillus subtilis glutaminase YbgJ and 5-oxo-L-norleucine formed by reaction of the protein with 6-diazo-5-oxo-L-norleucine | | Descriptor: | 5-OXO-L-NORLEUCINE, Glutaminase 1 | | Authors: | Singer, A.U, Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
1SAX
 
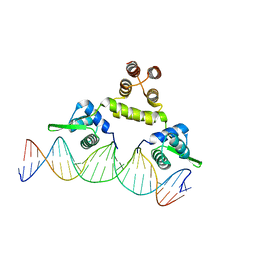 | | Three-dimensional structure of s.aureus methicillin-resistance regulating transcriptional repressor meci in complex with 25-bp ds-DNA | | Descriptor: | 5'-d(CAAAATTACAACTGTAATATCGGAG)-3', 5'-d(GCTCCGATATTACAGTTGTAATTTT)-3', Methicillin resistance regulatory protein mecI, ... | | Authors: | Garcia-Castellanos, R, Mallorqui-Fernandez, G, Marrero, A, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the transcriptional regulation of methicillin resistance: MecI repressor in complex with its operator
J.Biol.Chem., 279, 2004
|
|
1XP4
 
 | | Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, IODIDE ION, SULFATE ION | | Authors: | Morlot, C, Pernot, L, Le Gouellec, A, Di Guilmi, A.M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae
J.Biol.Chem., 280, 2005
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
6G5S
 
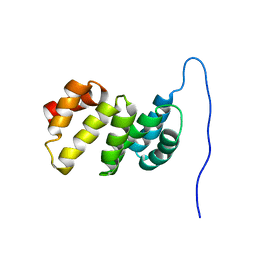 | | Solution structure of the TPR domain of the cell division coordinator, CpoB | | Descriptor: | Cell division coordinator CpoB | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Vollmer, W, Egan, A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
1ESI
 
 | |
4RYE
 
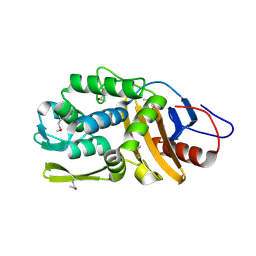 | | The crystal structure of D-ALANYL-D-ALANINE CARBOXYPEPTIDASE from Mycobacterium tuberculosis H37Rv | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Cuff, M, Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-12-15 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The crystal structure of D-ALANYL-D-ALANINE CARBOXYPEPTIDASE from Mycobacterium tuberculosis H37Rv
To be Published
|
|
1ES5
 
 | |
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | Descriptor: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | Deposit date: | 2020-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
4JMX
 
 | | Structure of LD transpeptidase LdtMt1 in complex with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Probable L,D-transpeptidase LdtA | | Authors: | Correale, S, Ruggiero, A, Capparelli, R, Pedone, E, Berisio, R. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of free and inhibited forms of the L,D-transpeptidase LdtMt1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JMN
 
 | | Crystal structure of LD transpeptidase LdtMt1 from M. tuberculosis | | Descriptor: | Probable L,D-transpeptidase LdtA | | Authors: | Ruggiero, A, Correale, S, Berisio, R. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of free and inhibited forms of the L,D-transpeptidase LdtMt1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1W7F
 
 | |
6NTW
 
 | | Crystal structure of E. coli YcbB | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable L,D-transpeptidase YcbB, SULFATE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
8P1U
 
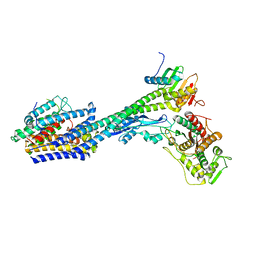 | | Structure of divisome complex FtsWIQLB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Yang, L, Chang, S, Tang, D, Dong, H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
2VGK
 
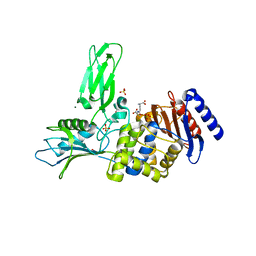 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
2VGJ
 
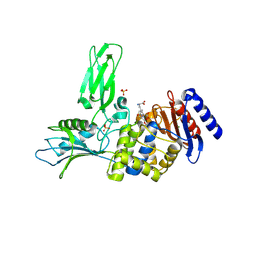 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | CEPHALOSPORIN, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
1BLC
 
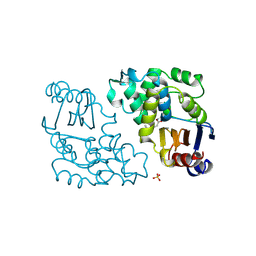 | |
