6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
2LBI
 
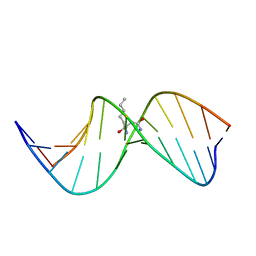 | | N2-dG:N2-dG interstrand cross-link induced by trans-4-hydroxynonenal | | Descriptor: | (4S)-nonane-1,4-diol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*CP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Kozekov, I.D, Wang, H, Kozekova, A, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Formation of a N2-dG:N2-dG carbinolamine DNA cross-link by the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N2-dG adduct.
J.Am.Chem.Soc., 133, 2011
|
|
2NC1
 
 | |
2R0K
 
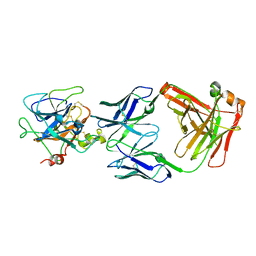 | | Protease domain of HGFA with inhibitor Fab58 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain of Fab58, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6OLP
 
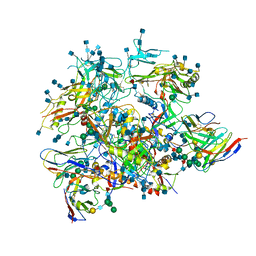 | | Full length HIV-1 Env AMC011 in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rantalainen, K, Cottrell, C.A. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
3SL9
 
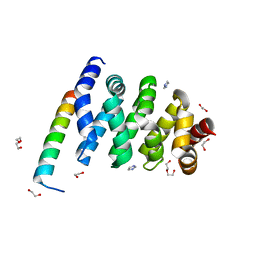 | | X-ray structure of Beta catenin in complex with Bcl9 | | Descriptor: | 1,2-ETHANEDIOL, B-cell CLL/lymphoma 9 protein, Catenin beta-1, ... | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
3SLA
 
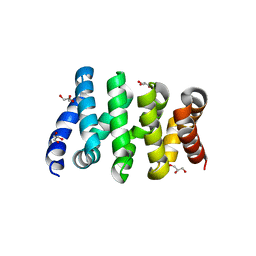 | | X-ray structure of first four repeats of human beta-catenin | | Descriptor: | Catenin beta-1, GLYCEROL, SODIUM ION | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
5FUU
 
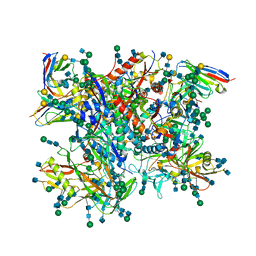 | | Ectodomain of cleaved wild type JR-FL EnvdCT trimer in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Em Structure of a Native, Fully Glycosylated and Cleaved HIV-1 Envelope Trimer
Science, 351, 2016
|
|
1UOR
 
 | |
6UR5
 
 | |
6WCR
 
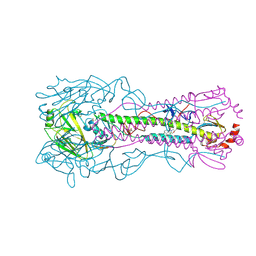 | |
7L56
 
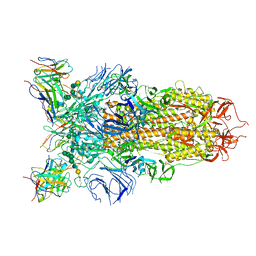 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7CUV
 
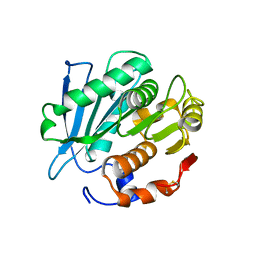 | |
7E30
 
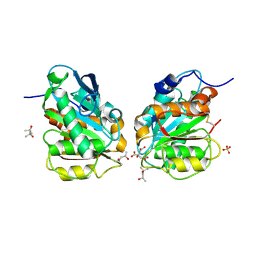 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E31
 
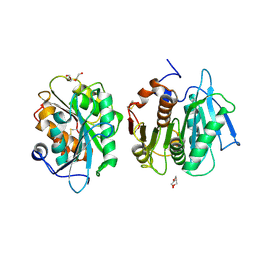 | | Crystal structure of a novel alpha/beta hydrolase mutant in apo form | | Descriptor: | TRIETHYLENE GLYCOL, alpha/beta hydrolase | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
6KBM
 
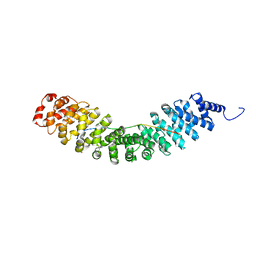 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6KGX
 
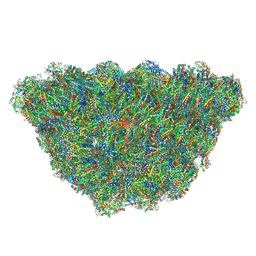 | | Structure of the phycobilisome from the red alga Porphyridium purpureum | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | Authors: | Sui, S.F, Ma, J.F, You, X, Sun, S. | | Deposit date: | 2019-07-12 | | Release date: | 2019-12-11 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of energy transfer in Porphyridium purpureum phycobilisome.
Nature, 579, 2020
|
|
1XYD
 
 | | NMR Solution Structure of Rat Zinc-Calcium-S100B, 20 Structures | | Descriptor: | CALCIUM ION, S-100 protein, beta chain, ... | | Authors: | Wilder, P.T, Varney, K.M, Weber, D.J. | | Deposit date: | 2004-11-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Zinc- and Calcium-Bound Rat S100B as Determined by Nuclear Magnetic Resonance Spectroscopy
Biochemistry, 44, 2005
|
|
5T3Z
 
 | |
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
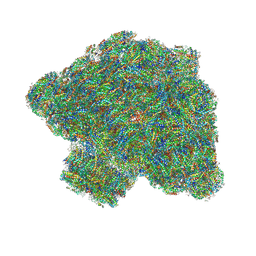 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
2HLI
 
 | | Solution structure of Crotonaldehyde-Derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct in the 5'-CpG-3' Sequence | | Descriptor: | DNA dodecamer, DNA dodecamer with S-crotonaldehyde adduct | | Authors: | Cho, Y.-J, Wang, H, Kozekov, I.D, Kurtz, A.J, Jacob, J, Voehler, M, Smith, J, Harris, T.M, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Orientation of the Crotonaldehyde-Derived N(2)-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct Hinders Interstrand Cross-Link Formation in the 5'-CpG-3' Sequence
Chem.Res.Toxicol., 19, 2006
|
|
1AGO
 
 | |
7AQ1
 
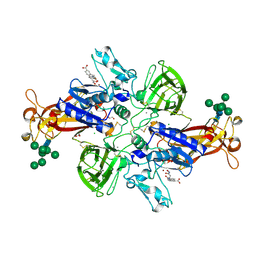 | | Crystal structure of human mature meprin beta in complex with the specific inhibitor MWT-S-270 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Linnert, M, Parthier, C, Fritz, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure and Dynamics of Meprin beta in Complex with a Hydroxamate-Based Inhibitor.
Int J Mol Sci, 22, 2021
|
|
5COD
 
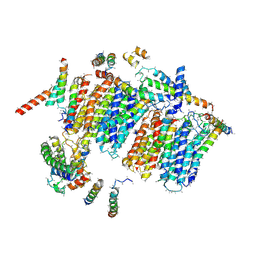 | | Bovine heart complex I membrane domain | | Descriptor: | NADH-ubiquinone oxidoreductase chain 4, NADH-ubiquinone oxidoreductase chain 5, SDAP, ... | | Authors: | Zhu, J, Hirst, J, King, M.S, Yu, M, Leslie, A.G.W, Klipcan, L. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.74 Å) | | Cite: | Structure of subcomplex I beta of mammalian respiratory complex I leads to new supernumerary subunit assignments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
