3O78
 
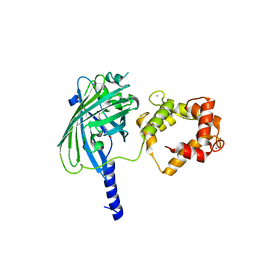 | | The structure of Ca2+ Sensor (Case-12) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, smooth muscle,Green fluorescent protein,Green fluorescent protein,Calmodulin-1 | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
2ZMW
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 6.0 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
7XSU
 
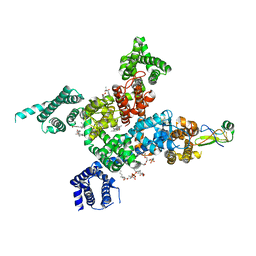 | | Cardiac sodium channel in complex with LqhIII | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-like toxin Lqh3, ... | | Authors: | Jiang, D, Catterall, W.A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for Nav1.5 Opening Modulated by a Gating Modifier Toxin
To Be Published
|
|
2YFP
 
 | | STRUCTURE OF YELLOW-EMISSION VARIANT OF GFP | | Descriptor: | PROTEIN (GREEN FLUORESCENT PROTEIN) | | Authors: | Wachter, R.M, Elsliger, M.A, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein.
Structure, 6, 1998
|
|
1YZW
 
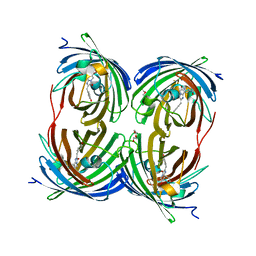 | | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore | | Descriptor: | DI(HYDROXYETHYL)ETHER, GFP-like non-fluorescent chromoprotein | | Authors: | Wilmann, P.G, Petersen, J, Pettikiriarachchi, A, Buckle, A.M, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore
J.Mol.Biol., 349, 2005
|
|
6WSG
 
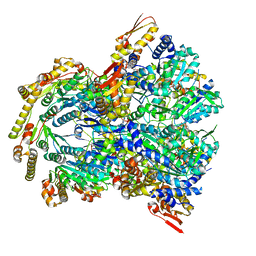 | | ClpX-ClpP complex bound to ssrA-tagged GFP, intermediate complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, Green fluorescent protein, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
8B6T
 
 | | X-ray structure of the interface optimized haloalkane dehalogenase HaloTag7 fusion to the green fluorescent protein GFP (ChemoG5-TMR) labeled with a chloroalkane tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, Green fluorescent protein,Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
8B6S
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 fusion to the green fluorescent protein GFP (ChemoG1) labeled with a chloroalkane tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein,Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
7FBS
 
 | | structure of a channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-[2-[(2R)-2-oxidanyl-3-(propylamino)propoxy]phenyl]-3-phenyl-propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.J, Catterall, W.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Open-state structure and pore gating mechanism of the cardiac sodium channel.
Cell, 184, 2021
|
|
6WRF
 
 | | ClpX-ClpP complex bound to GFP-ssrA, recognition complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
6WVF
 
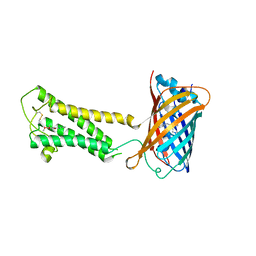 | | E.coli DsbB C104S with ubiquinone | | Descriptor: | Green fluorescent protein,Disulfide bond formation protein B,Green fluorescent protein, UBIQUINONE-1 | | Authors: | Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
6WVE
 
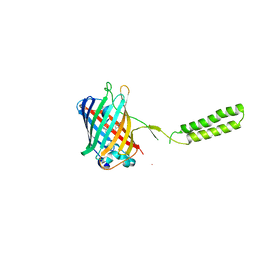 | | Chicken SPCS1 | | Descriptor: | Green fluorescent protein,Chicken Signal Peptidase Complex Subunit 1,Green fluorescent protein | | Authors: | Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
8ET3
 
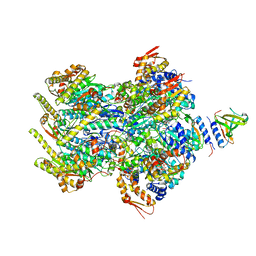 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y96
 
 | |
2GX2
 
 | | Crystal structural and functional analysis of GFP-like fluorescent protein Dronpa | | Descriptor: | MAGNESIUM ION, fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.-H, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the photoswitchable fluorescent protein Dronpa-C62S
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2GX0
 
 | |
2A54
 
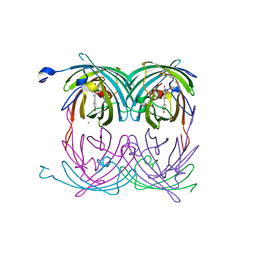 | | fluorescent protein asFP595, A143S, on-state, 1min irradiation | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A53
 
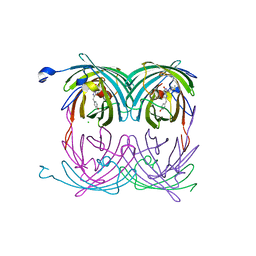 | | fluorescent protein asFP595, A143S, off-state | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6UQE
 
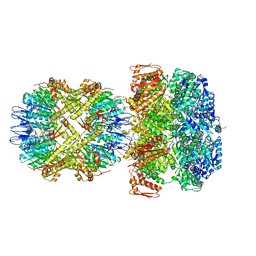 | | ClpA/ClpP Disengaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Lopez, K.L, Rizo, A.R, Southworth, D.R. | | Deposit date: | 2019-10-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3VHT
 
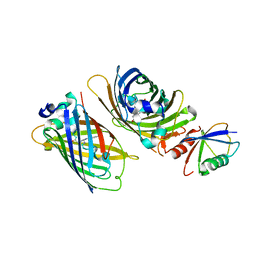 | | Crystal structure of GFP-Wrnip1 UBZ domain fusion protein in complex with ubiquitin | | Descriptor: | Green fluorescent protein, Green fluorescent protein,ATPase WRNIP1, Ubiquitin, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2011-09-06 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 2016
|
|
6UQO
 
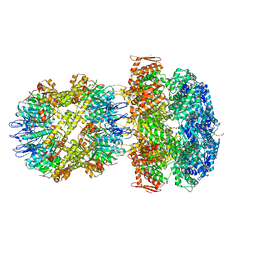 | | ClpA/ClpP Engaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp endopeptidase proteolytic subunit ClpP, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Southworth, D.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2Q6P
 
 | | The Chemical Control of Protein Folding: Engineering a Superfolder Green Fluorescent Protein | | Descriptor: | Green fluorescent protein mutant 3 | | Authors: | Steiner, T, Hess, P, Bae, J.H, Wiltschi, B, Moroder, L, Budisa, N. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthetic biology of proteins: tuning GFPs folding and stability with fluoroproline.
Plos One, 3, 2008
|
|
4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPK
 
 | | Crystal structure of eCGP123 T69V variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
3VIC
 
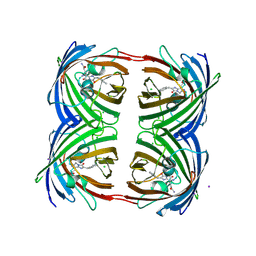 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
