5GTK
 
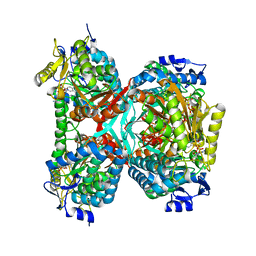 | | NAD+ complex structure of aldehyde dehydrogenase from bacillus cereus | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Ngo, H.P.T, Hong, S.H, Ho, T.H, Oh, D.K, Kang, L.W. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of aldehyde dehydrogenase from Bacillus cereus having atypical bidirectional oxidizing and reducing activities for all-trans-retinal
To Be Published
|
|
9EZ0
 
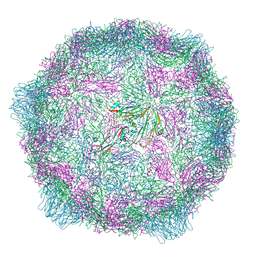 | | Poliovirus type 1 (strain Mahoney) expanded conformation stabilised virus-like particle (PV1 SC6b) from a yeast expression system. | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Bahar, M.W, Sherry, L, Stonehouse, N.J, Rowlands, D.J, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-10 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
3U9W
 
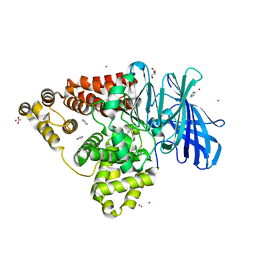 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor sc57461A | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Niegowski, D, Thunnissen, M, Tholander, F, Rinaldo-Matthis, A, Muroya, A, Haeggstrom, J.Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of human Leukotriene A4 hydrolase in complex with inhibitor sc57461A
To be Published
|
|
4YSV
 
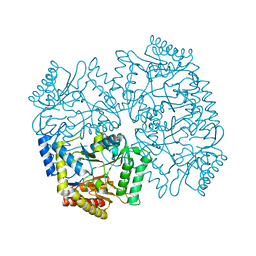 | |
1RFX
 
 | | Crystal Structure of resisitin | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
8G45
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
4YL6
 
 | |
5CZO
 
 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 2 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
8G43
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8Z48
 
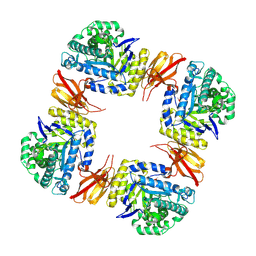 | | Beta-galactosidase from Bacteroides xylanisolvens (complex with methyl beta-galactopyranose) | | Descriptor: | Beta-galactosidase, methyl beta-D-galactopyranoside | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
5CT3
 
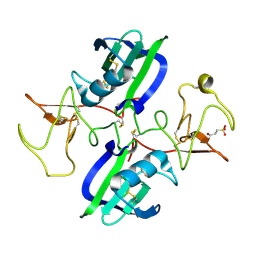 | | The structure of the NK1 fragment of HGF/SF complexed with 2FA | | Descriptor: | 3-hydroxypropane-1-sulfonic acid, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
4IN9
 
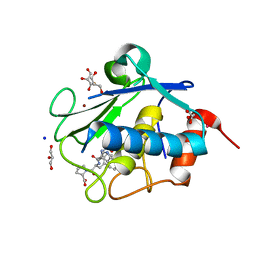 | | Structure of karilysin MMP-like catalytic domain in complex with inhibitory tetrapeptide SWFP | | Descriptor: | GLYCEROL, Karilysin protease, POTASSIUM ION, ... | | Authors: | Guevara, T, Ksiazek, M, Skottrup, P.D, Cerda-Costa, N, Trillo-Muyo, S, de Diego, I, Riise, E, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the catalytic domain of the Tannerella forsythia matrix metallopeptidase karilysin in complex with a tetrapeptidic inhibitor.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5U9X
 
 | | Ocellatin-LB2 | | Descriptor: | Ocellatin-K1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-LB2
To Be Published
|
|
8Z47
 
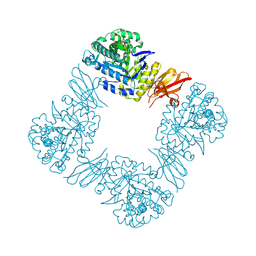 | | Beta-galactosidase from Bacteroides xylanisolvens (ligand-free) | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Nakajima, M, Motouchi, S, Kobayashi, K. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and function of a beta-1,2-galactosidase from Bacteroides xylanisolvens, an intestinal bacterium.
Commun Biol, 8, 2025
|
|
6MAU
 
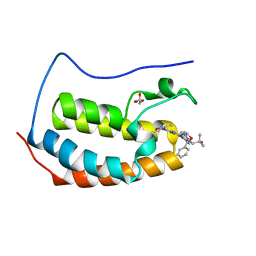 | | Crystal structure of human BRD4(1) in complex with CN210 (compound 19) | | Descriptor: | 1-(4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-[(3S)-3-phenylmorpholin-4-yl]quinazolin-2-yl}-1H-pyrazol-1-yl)-2-methylpropan-2-ol, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Nadupalli, A, Fontano, E, Connors, C.R, Chan, S.G, Olland, A.M, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Lead optimization and efficacy evaluation of quinazoline-based BET family inhibitors for potential treatment of cancer and inflammatory diseases.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5U02
 
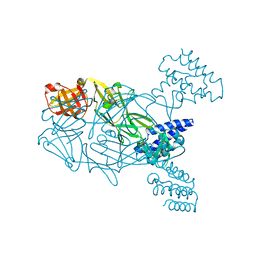 | | Crystal structure of S. aureus TarS 217-571 | | Descriptor: | Glycosyl transferase, IMIDAZOLE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-22 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
4BYZ
 
 | | Structural characterization using Sulfur-SAD of the cytoplasmic domain of Burkholderia pseudomallei PilO2Bp, an actin-like protein component of a Type IVb R64-derivative pilus machinery. | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, TYPE IV PILUS BIOSYNTHESIS PROTEIN | | Authors: | Lassaux, P, Manjasetty, B.A, Conchillo-Sole, O, Yero, D, Gourlay, L, Perletti, L, Daura, X, Belrhali, H, Bolognesi, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Redefining the Pf06864 Pfam Family Based on Burkholderia Pseudomallei Pilo2BP S-Sad Crystal Structure.
Plos One, 9, 2014
|
|
8G44
 
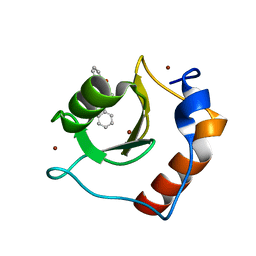 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
4C90
 
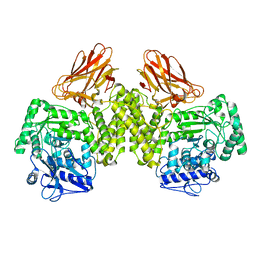 | | Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility | | Descriptor: | ALPHA-GLUCURONIDASE GH115, SODIUM ION | | Authors: | Rogowski, A, Basle, A, Farinas, C.S, Solovyova, A, Mortimer, J.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evidence that Gh115 Alpha-Glucuronidase Activity is Dependent on Conformational Flexibility
J.Biol.Chem., 289, 2014
|
|
4IP1
 
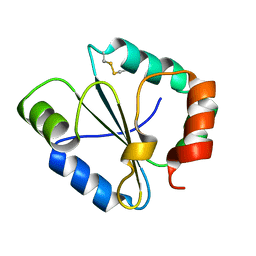 | |
6MBQ
 
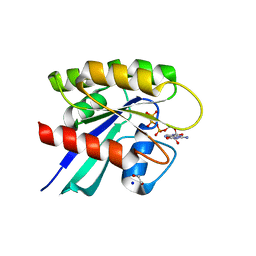 | | Crystal structure of Mg-free wild-type KRAS (2-166) bound to GMPPNP in the state 1 conformation | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dharmaiah, S, Davies, D.R, Abendroth, J, Gillette, W.G, Stephen, A.G, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
5QC5
 
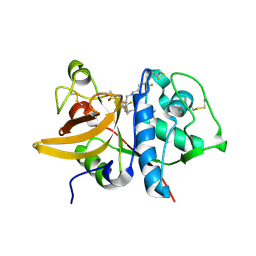 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[5-{1-[3-(4-tert-butylpiperidin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}-2-(trifluoromethyl)phenyl]-N-[(4-fluorophenyl)methyl]methanamine, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
4YQF
 
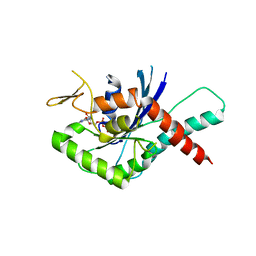 | | GTPase domain of Human Septin 9 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-9 | | Authors: | Matos, S.O, Leonardo, D.A, Macedo, J.N, Pereira, H.M, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | GTPase domain of Human Septin 9
To Be Published
|
|
4IRA
 
 | | CobR in complex with FAD | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Lawrence, A.D, Scott, A.F, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biophysical characterisation and structure-function analysis of Brucella melitensis CobR: Protein-flavin interactions determine function and stability
To be Published
|
|
6M9B
 
 | |
