3ZGQ
 
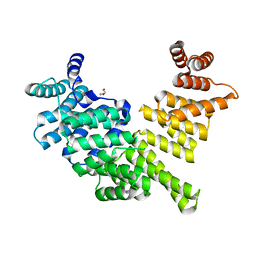 | | Crystal structure of human interferon-induced protein IFIT5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, INTERFERON-INDUCED PROTEIN WITH TETRATRICOPEPTIDE REPEATS 5 | | Authors: | Katibah, G.E, Lee, H.J, Huizar, J.P, Vogan, J.M, Alber, T, Collins, K. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-23 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | TRNA Binding, Structure, and Localization of the Human Interferon-Induced Protein Ifit5.
Mol.Cell, 49, 2013
|
|
2GWW
 
 | |
2FH2
 
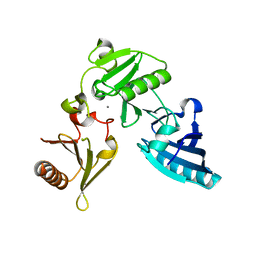 | | C-terminal half of gelsolin soaked in EGTA at pH 4.5 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
2FH4
 
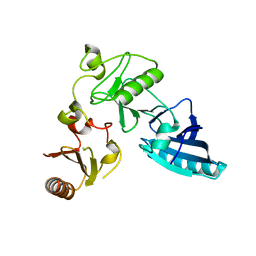 | | C-terminal half of gelsolin soaked in EGTA at pH 8 | | Descriptor: | Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
2FH1
 
 | | C-terminal half of gelsolin soaked in low calcium at pH 4.5 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
7AA8
 
 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
5M05
 
 | | Chicken smooth muscle myosin motor domain co-crystallized with the specific CK-571 inhibitor, MgADP form | | Descriptor: | 4-{[(2-chloro-3-fluorobenzyl)carbamoyl](methyl)amino}-3,4-dideoxy-5-O-(isoquinolin-3-ylcarbamoyl)-D-erythro-pentitol, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sirigu, S, Hartman, J, Houdusse, A. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | Highly selective inhibition of myosin motors provides the basis of potential therapeutic application.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5N69
 
 | | Cardiac muscle myosin S1 fragment in the pre-powerstroke state co-crystallized with the activator Omecamtiv Mecarbil | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Planelles-Herrero, V.J, Hartman, J.J, Robert-Paganin, J, Malik, F.I, Houdusse, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun, 8, 2017
|
|
7BV4
 
 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N9C
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-OH | | Descriptor: | Ac-[2-Cl-F]-PP-[ProM-1]-OH, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NBF
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-3]-OH | | Descriptor: | (3~{S},6~{S},7~{R},9~{a}~{R})-6-[[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonylamino]-7-methyl-5-oxidanylidene-1,2,3,6,7,9~{a}-hexahydropyrrolo[1,2-a]azepine-3-carboxylic acid, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NAJ
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-1]-[ProM-1]-OH | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{R},7~{S},10~{S},13~{R})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{S},7~{R},10~{R},13~{S})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, CHLORIDE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NCF
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-4]-OH | | Descriptor: | (1~{S},4~{S},7~{R},10~{R})-14-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-3,14-diazatricyclo[8.4.0.0^{3,7}]tetradec-8-ene-4-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N9P
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-NH2 | | Descriptor: | Ac-[2-Cl-F]-PP-[ProM-1]-NH2, CHLORIDE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N91
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPP-OH | | Descriptor: | Ac-[2-Cl-F]-PPPP-OH, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N6A
 
 | | Cardiac muscle myosin motor domain in the pre-powerstroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Planelles-Herrero, V.J, Hartman, J.J, Robert-Paganin, J, Malik, F.I, Houdusse, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun, 8, 2017
|
|
5NCG
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-9]-OH | | Descriptor: | (3~{S},7~{R},10~{R},11~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-11-methyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-04 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2YVC
 
 | |
2H7V
 
 | | Co-crystal structure of YpkA-Rac1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Migration-inducing protein 5, ... | | Authors: | Prehna, G, Ivanov, M, Bliska, J.B, Stebbins, C.E. | | Deposit date: | 2006-06-04 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Yersinia virulence depends on mimicry of host rho-family nucleotide dissociation inhibitors.
Cell(Cambridge,Mass.), 126, 2006
|
|
3D2U
 
 | | Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Actin, Beta-2-microglobulin, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2008-05-08 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2LJ8
 
 | |
6HYN
 
 | | Structure of ATG13 LIR motif bound to GABARAP | | Descriptor: | Autophagy-related protein 13,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HOG
 
 | | Structure of VPS34 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HYO
 
 | | Structure of ULK1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
7EA7
 
 | | crystal structure of NAP1 LIR in complex with GABARAP | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, NAP1_LIR motif | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
