6GK6
 
 | |
5DQY
 
 | | A fully oxidized human thioredoxin | | Descriptor: | BENZOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hwang, J. | | Deposit date: | 2015-09-15 | | Release date: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of fully oxidized human thioredoxin.
Biochem.Biophys.Res.Commun., 467, 2015
|
|
6GIN
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with an Quinazolinone based ALK2 inhibitor with a 4-morpholinophenyl solvent accessible group. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-morpholin-4-ylphenyl)-6-quinolin-4-yl-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Kopec, J, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
6GJ8
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH BI 2852 | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5E3J
 
 | | The response regulator RstA is a potential drug target for Acinetobacter baumannii | | Descriptor: | Response regulator RstA | | Authors: | Russo, T.A, Manohar, A, Beanan, J.M, Olson, R, MacDonald, U, Graham, J, Umland, T.C. | | Deposit date: | 2015-10-02 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Response Regulator BfmR Is a Potential Drug Target for Acinetobacter baumannii.
Msphere, 1, 2016
|
|
5DR3
 
 | | Endothiapepsin in complex with fragment 333 | | Descriptor: | 1,2-ETHANEDIOL, 4-propan-2-ylsulfanyl-1-propyl-6,7-dihydro-5~{H}-cyclopenta[d]pyrimidin-2-one, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6UKC
 
 | |
5DRD
 
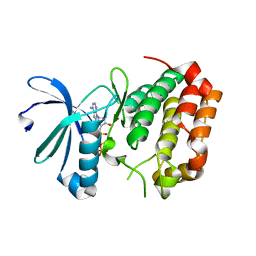 | | Aurora A Kinase in Complex with ATP in Space Group P6122 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, MAGNESIUM ION | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
3DOX
 
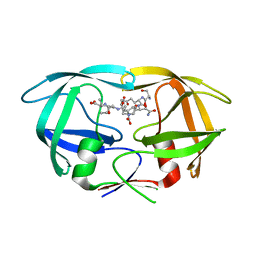 | | X-ray structure of HIV-1 protease in situ product complex | | Descriptor: | A PEPTIDE SUBSTRATE-PIV, A PEPTIDE SUBSTRATE-SQNY, HIV-1 PROTEASE | | Authors: | Hosur, M.V, Ferrer, J.-L, Das, A, Prashar, V, Bihani, S. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of HIV-1 protease in situ product complex
Proteins, 74, 2009
|
|
6UK1
 
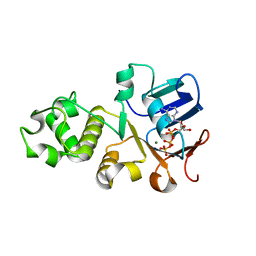 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
6GOB
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, Lysozyme C, N,N-pyridylbenzimidazole derivative-Pd complex, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6GOJ
 
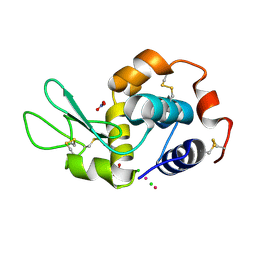 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated triphenylphosphonium cation | | Descriptor: | CHLORIDE ION, Lysozyme C, NITRATE ION, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
5E7Y
 
 | | Crystal structure of P450 BM3 heme domain M7 variant | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-13 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of P450 BM3 heme domain M7 variant
To Be Published
|
|
3DUT
 
 | | The high salt (phosphate) crystal structure of deoxy hemoglobin E (GLU26LYS) at physiological pH (pH 7.35) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2008-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The high salt (phosphate) crystal structure of deoxy
hemoglobin E (GLU26LYS) at physiological pH (pH 7.35)
To be Published
|
|
8B78
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{R})-8-(2-chloranyl-6-oxidanyl-phenyl)-7-fluoranyl-9-prop-1-ynyl-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
6USZ
 
 | | Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vigers, G.P, Smith, D.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the Clinical Development CandidateMRTX849, a Covalent KRASG12CInhibitor for the Treatment of Cancer.
J.Med.Chem., 63, 2020
|
|
3DRM
 
 | | 2.2 Angstrom Crystal Structure of Thr114Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
5E8T
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) | | Descriptor: | GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6GQZ
 
 | | Petrobactin-binding engineered lipocalin without ligand | | Descriptor: | Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2018-06-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reprogramming Human Siderocalin To Neutralize Petrobactin, the Essential Iron Scavenger of Anthrax Bacillus.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5E90
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F, S280T,Y282F,S287N,A350C,L352F) IN COMPLEX WITH 3-AMINO-6- [4-(2-HYDROXYETHYL)PHENYL]-N-[4-(MORPHOLIN-4-YL)PYRIDIN-3-YL]PYRAZINE-2-CARBOXAMIDE | | Descriptor: | 3-amino-6-[4-(2-hydroxyethyl)phenyl]-N-[4-(morpholin-4-yl)pyridin-3-yl]pyrazine-2-carboxamide, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3DS6
 
 | | P38 complex with a phthalazine inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Grosfeld, D. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
3DIC
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V108A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
6UPJ
 
 | | HIV-2 PROTEASE/U99294 COMPLEX | | Descriptor: | 6,7,8,9-TETRAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOHEPTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
8BVS
 
 | | Cryo-EM structure of rat SLC22A6 bound to tenofovir | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody), ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
