7SVO
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
6ZR9
 
 | |
7ZQW
 
 | | Structure of the SARS-CoV-1 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Muriel-Goni, S, Fabrega-Ferrer, M, Herrera-Morande, A, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
6VC2
 
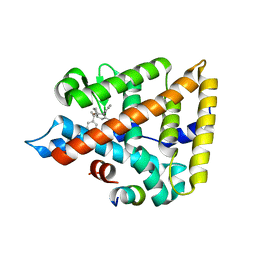 | | LRH-1 bound to SS-RJW100 and a fragment of the Tif2 Coactivator | | Descriptor: | (1S,3aS,6aS)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Enantiomer-specific activities of an LRH-1 and SF-1 dual agonist.
Sci Rep, 10, 2020
|
|
7TC3
 
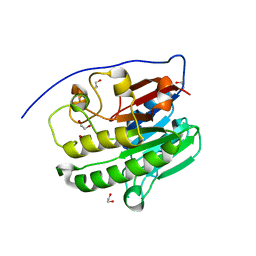 | | Human APE1 in the apo form | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
6VKK
 
 | |
5WCK
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
6N3J
 
 | |
8A9C
 
 | |
7TXG
 
 | | Structure of the Class II Fructose-1,6-Bisphosphatase from Francisella tularensis with native Mn++ divalent cation and partially occupied product F6P | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Abad-Zapatero, C, Selezneva, A.I, Harding, L.N.M, Movahedzadeh, F. | | Deposit date: | 2022-02-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New structures of Class II Fructose-1,6-Bisphosphatase from Francisella tularensis provide a framework for a novel catalytic mechanism for the entire class.
Plos One, 18, 2023
|
|
5WDF
 
 | |
5WB9
 
 | | Crystal structure of CD4 binding site antibody N60P23 in complex with HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Gohain, N, Tolbert, W, Pazgier, M. | | Deposit date: | 2017-06-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Near-Pan-neutralizing Antibodies against HIV-1 by Deconvolution of Plasma Humoral Responses.
Cell, 173, 2018
|
|
9FZQ
 
 | | Proton conductance by human uncoupling protein 1 is inhibited by both purine and pyrimidine nucleotides | | Descriptor: | CA9865, CA9871, CARDIOLIPIN, ... | | Authors: | Jones, S.A, Sowton, A.P, Lacabanne, D, King, M.S, Palmer, S.M, Zogg, T, Pardon, E, Steyaert, J, Ruprecht, J.J, Kunji, E.R.S. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Proton conductance by human uncoupling protein 1 is inhibited by purine and pyrimidine nucleotides.
Embo J., 44, 2025
|
|
6N3U
 
 | |
5K72
 
 | | IRAK4 in complex with Compound 21 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5JTS
 
 | | Structure of a beta-1,4-mannanase, SsGH134. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
7BP4
 
 | |
5K7G
 
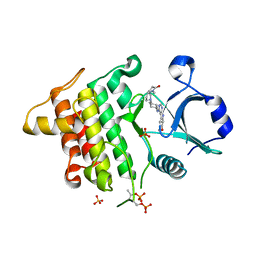 | | IRAK4 in complex with AZ3862 | | Descriptor: | (3~{a}~{S},7~{a}~{R})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
8AV9
 
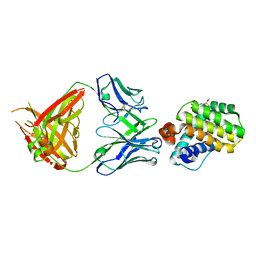 | | INDUCED MYELOID LEUKEMIA CELL DIFFERENTIATION PROTEIN FABCOMPLEX IN COMPLEX WITH COMPOUND 1 | | Descriptor: | (3R,6R,7S,8E,11S,12R,22S)-6'-chloro-7-methoxy-11,12-dimethyl-13,13-dioxo-spiro[20-oxa-13-gamma6-thia-1,14-diazatetracyclo[14.7.2.03,6.019,24]pentacosa-8,16(25),17,19(24)-tetraene-22,1'-tetralin]-15-one, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2022-08-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Design of rigid protein-protein interaction inhibitors enables targeting of undruggable Mcl-1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6VUC
 
 | |
7BP2
 
 | |
5X08
 
 | | Crystal structure of broadly neutralizing anti-HIV-1 antibody 4E10, mutant Npro, with peptide bound | | Descriptor: | ACETATE ION, CHLORIDE ION, Envelope glycoprotein gp160, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Functional Contacts between MPER and the Anti-HIV-1 Broadly Neutralizing Antibody 4E10 Extend into the Core of the Membrane
J. Mol. Biol., 429, 2017
|
|
5JRM
 
 | | Crystal Structure of a Xylanase at 1.56 Angstroem resolution | | Descriptor: | Endo-1,4-beta-xylanase, GLYCEROL, SULFATE ION | | Authors: | Gomez, S, Payne, A.M, Savko, M, Fox, G.C, Shepard, W.E, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Biotechnol Biofuels, 9, 2016
|
|
6YBC
 
 | | RNASE 3/1 version2 phosphate complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNASE 3/1 version2 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-03-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
