8DSF
 
 | | Structure of cIAP1 with BCCov | | Descriptor: | (4S)-4-[2-(2-{4-[(2E)-4-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}-4-oxobut-2-en-1-yl]piperazin-1-yl}ethoxy)acetamido]-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-L-prolinamide unbound form, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Schiemer, J.S, Calabrese, M.F. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A covalent BTK ternary complex compatible with targeted protein degradation.
Nat Commun, 14, 2023
|
|
6XYG
 
 | | Femtosecond structure of bovine trypsin at room temperature | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Jensen, M. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.50000238 Å) | | Cite: | High-resolution macromolecular crystallography at the FemtoMAX beamline with time-over-threshold photon detection
J.Synchrotron Radiat., 2021
|
|
6S4T
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 2-[4-[[3-[3-(phenylmethyl)-8-(trifluoromethyl)quinolin-4-yl]phenoxy]methyl]phenyl]ethanoic acid, Oxysterols receptor LXR-beta | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S5K
 
 | | LXRbeta ligand binding domain in complex with small molecule inhibitors | | Descriptor: | 3-(4-phenylbutylamino)-1,4-bis(phenylmethyl)pyrrole-2,5-dione, Oxysterols receptor LXR-beta | | Authors: | Petersen, J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S4U
 
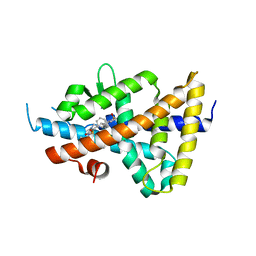 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 6-[4-[[3-oxidanyl-1,1-bis(oxidanylidene)-5-phenyl-2-propan-2-yl-3~{H}-1,2-thiazol-4-yl]amino]butyl]pyridine-2-sulfonamide, Oxysterols receptor LXR-beta | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S4N
 
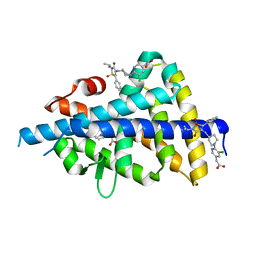 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 2-[5-chloranyl-6-[4-[[1,1,3-tris(oxidanylidene)-5-phenyl-2-propan-2-yl-1,2-thiazol-4-yl]amino]piperidin-1-yl]pyridin-3-yl]ethanoic acid, Oxysterols receptor LXR-beta, SULFATE ION | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
4TX2
 
 | |
8G1R
 
 | |
6CUC
 
 | |
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
1LU0
 
 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7UNX
 
 | | NMR solution structure of xanthusin-1 | | Descriptor: | Xanthusin-1 | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Discovery of Five Classes of Bacterial Defensins: Ancestral Precursors of Defensins from Eukarya?
Acs Omega, 2024
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGW
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3- HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE | | Descriptor: | 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5QII
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-(3-(1-(4-CHLOROPHENYL)CYCLOPROPYL) -[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8-YL)PROPAN-2-OL | | Descriptor: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
6ZWK
 
 | | Crystal structure of the phosphorylated C-terminal tail of histone H2AX in complex with a specific nanobody (C6 gammaXbody) | | Descriptor: | CHLORIDE ION, Histone H2AX, SODIUM ION, ... | | Authors: | McEwen, A.G, Moeglin, E, Desplancq, D, Weiss, E, Poterszman, A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Novel Nanobody Precisely Visualizes Phosphorylated Histone H2AX in Living Cancer Cells under Drug-Induced Replication Stress.
Cancers (Basel), 13, 2021
|
|
5QCM
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[[(1~{S})-2-[(~{E})-3-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]phenyl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCL
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
8JDI
 
 | |
8JDH
 
 | |
8TB0
 
 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | Descriptor: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TB7
 
 | | Cryo-EM Structure of GPR61- | | Descriptor: | 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide, Fab hinge-binding nanobody, Fab24 BAK5 heavy chain, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TOQ
 
 | | ACE2-peptide 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOS
 
 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOR
 
 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
