6G30
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6VI3
 
 | | Straight Filament from Alzheimer's Disease Human Brain Tissue | | Descriptor: | GLYCINE, Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains
Cell, 180, 2020
|
|
5V3G
 
 | | PRDM9-allele-C ZnF8-13 | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*AP*AP*CP*GP*CP*TP*CP*AP*CP*TP*GP*GP*GP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*CP*CP*AP*GP*TP*GP*AP*GP*CP*GP*TP*TP*GP*CP*CP*C)-3'), PR domain zinc finger protein 9, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
J. Biol. Chem., 292, 2017
|
|
5V5C
 
 | | VQIINK, Structure of the amyloid-spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-14 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.25 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
5U24
 
 | | X-ray structure of the WlaRG aminotransferase from campylobacter jejuni, K184A mutant in complex with TDP-Fuc3N | | Descriptor: | (2R,3R,4S,5R,6R)-3,5-dihydroxy-4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U21
 
 | | X-ray structure of the WlaRF aminotransferase from Campylobacter jejuni, K184A mutant in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
6YW2
 
 | |
6G3U
 
 | | Structure of Pseudomonas aeruginosa Isocitrate Dehydrogenase, IDH | | Descriptor: | 2-OXOGLUTARIC ACID, Isocitrate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Crousilles, A, Welch, M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Gluconeogenic precursor availability regulates flux through the glyoxylate shunt inPseudomonas aeruginosa.
J. Biol. Chem., 293, 2018
|
|
6NMU
 
 | | Kick-Off Fab 115 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 115 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 115 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6FOE
 
 | | BaxB01 Fab fragment | | Descriptor: | Fab BaxB01 heavy chain, Fab BaxB01 light chain | | Authors: | Hollerweger, J, Schinagl, A, Kerschbaumer, R.J, Scheiflinger, F, Thiele, M, Goettig, P, Brandstetter, H. | | Deposit date: | 2018-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Biochemistry, 57, 2018
|
|
7K53
 
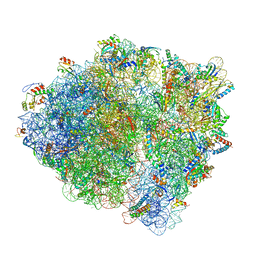 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
6VRA
 
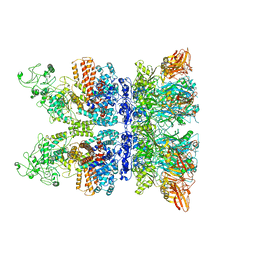 | | Anthrax octamer prechannel bound to full-length edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
6M8P
 
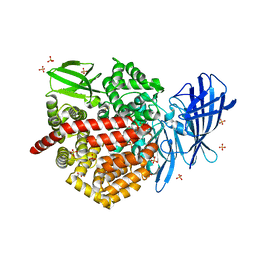 | | Human ERAP1 bound to phosphinic pseudotripeptide inhibitor DG013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, Nalpha-[(2S)-2-{[[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]methyl}-4-methylpentanoyl]-L-tryptophanamide, ... | | Authors: | Maben, Z, Stern, L.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Conformational dynamics linked to domain closure and substrate binding explain the ERAP1 allosteric regulation mechanism.
Nat Commun, 12, 2021
|
|
7UZN
 
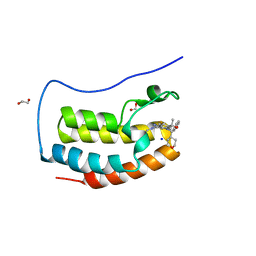 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMT-206059 AKA 2-{(3M)-3-(1,4-DIMETHYL-1H-1,2,3-TRIAZOL-5-YL)-8-FLUORO-5-[(S)-(OXAN-4-YL)(PHENYL)METHYL]-5H-PYRIDO[3,2-b]INDOL-7-YL}PROPAN-2-OL, TRIPLY DEUTERATED ON THE 4-METHYL GROUP | | Descriptor: | 1,2-ETHANEDIOL, 2-{(3M)-3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Development of BET Inhibitors as Potential Treatments for Cancer: Optimization of Pharmacokinetic Properties.
Acs Med.Chem.Lett., 13, 2022
|
|
5V5B
 
 | | KVQIINKKLD, Structure of the amyloid spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
7K73
 
 | |
6W6N
 
 | |
6G2V
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
5UIN
 
 | | X-ray structure of the W305A variant of the FdtF N-formyltransferase from salmonella enteric O60 | | Descriptor: | CHLORIDE ION, Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
7UQ3
 
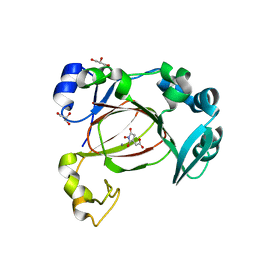 | | JmjC domain-containing protein 5 (JMJD5) in complex with Mn and (S)-2-(1-hydroxy-2,5-dioxopyrrolidin-3-yl)acetic acid | | Descriptor: | Bifunctional peptidase and arginyl-hydroxylase JMJD5, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2022-04-19 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.
Sci Rep, 12, 2022
|
|
5ULC
 
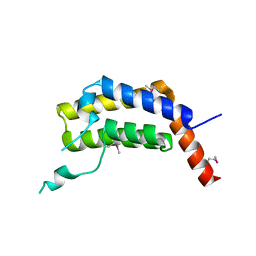 | | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328 | | Descriptor: | Bromodomain protein 1 | | Authors: | Wernimont, A.K, Amaya, M.F, Lam, A, Ali, A, Zhang, A.Z, Kenzina, L, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Hui, R, Walker, J.R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328
To be published
|
|
6YVX
 
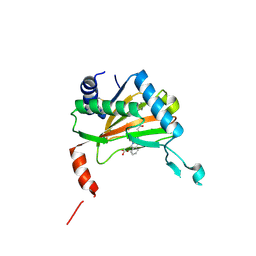 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic BB-287 | | Descriptor: | 4-(isoquinolin-3-ylamino)-4-oxobutanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
6YW4
 
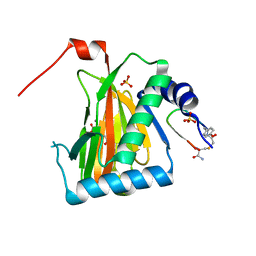 | |
7KP1
 
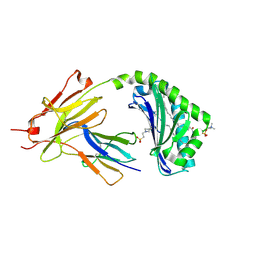 | | CD1a-42:2 SM binary complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, T-cell surface glycoprotein CD1a, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
6UE3
 
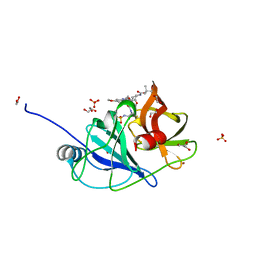 | |
