6MMB
 
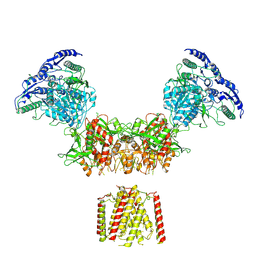 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Super-Splayed' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 6.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMS
 
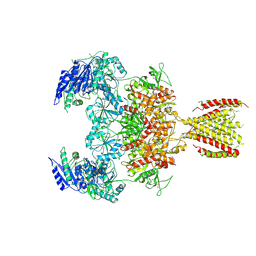 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar EDTA, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.38 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6V6T
 
 | |
6MGQ
 
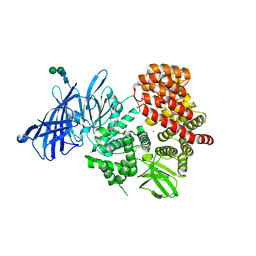 | | ERAP1 in the open conformation bound to 10mer phosphinic inhibitor DG014 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, Phosphinic inhibitor DG014, ... | | Authors: | Stern, L.J, Maben, Z. | | Deposit date: | 2018-09-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Conformational dynamics linked to domain closure and substrate binding explain the ERAP1 allosteric regulation mechanism.
Nat Commun, 12, 2021
|
|
6MHM
 
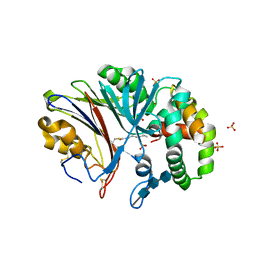 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dementiev, A, Joachimiak, A, Doan, N. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
6VB9
 
 | | Covalent adduct of cis-2,3-epoxysuccinic acid with Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Isocitrate lyase, ... | | Authors: | Krieger, I.V, Pham, T.V, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
6MMV
 
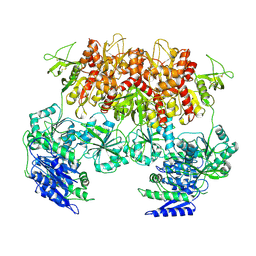 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* Extracellular Domain in the '2-Knuckle-Asymmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMN
 
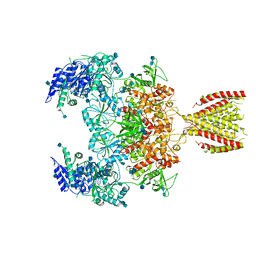 | | Diheteromeric NMDA receptor GluN1/GluN2A in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMG
 
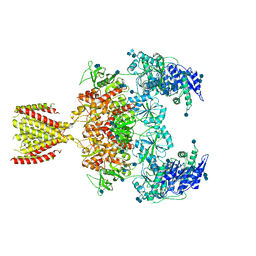 | | Diheteromeric NMDA receptor GluN1/GluN2A in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar EDTA, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.23 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMX
 
 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the 'Extended' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.99 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6YVW
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with monocyclic BB-328 | | Descriptor: | 4-[(5-bromanyl-4,6-dimethyl-pyridin-2-yl)amino]-4-oxidanylidene-butanoic acid, Egl nine homolog 1, FE (III) ION, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
6YW1
 
 | |
6YW3
 
 | |
6MHF
 
 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6YVT
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with MD-253 | | Descriptor: | 2-[[5-(6-methoxynaphthalen-2-yl)-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Egl nine homolog 1, GLYCEROL, ... | | Authors: | Chowdhury, R, Demetriades, M, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Dynamic combinatorial chemistry employing boronic acids/boronate esters leads to potent oxygenase inhibitors.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7JOQ
 
 | | Structure of NV1 small terminase | | Descriptor: | Small Terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
6YVZ
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic JLS-367 | | Descriptor: | 4-[(5-bromanylisoquinolin-3-yl)amino]-4-oxidanylidene-butanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Sorensen, J.L, Schofield, C.J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
6MMA
 
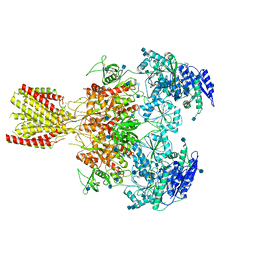 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Extended' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 6.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-29 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.31 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMT
 
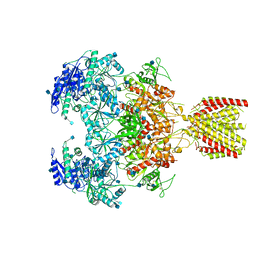 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '1-Knuckle' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.46 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MKN
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
6YW0
 
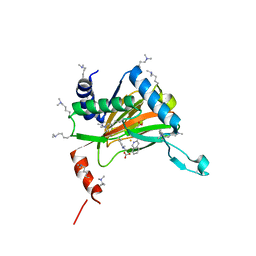 | |
5UIL
 
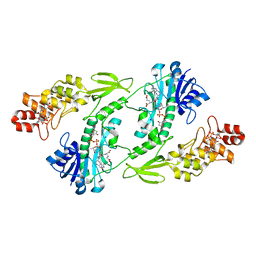 | | X-ray structure of the FdtF N-formyltransferase from Salmonella enterica O60 in complex with TDP-Fuc3N and tetrahydrofolate | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, Formyltransferase, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
6XVD
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3F) in pH7.4 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1-W3F | | Authors: | Xue, G.P, Xie, X, Zhou, Y, Yuan, C, Huang, M.D, Jiang, L.G. | | Deposit date: | 2020-01-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6FZR
 
 | | Crystal structure of scFv-SM3 in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-[(fluoroacetyl)amino]-alpha-D-galactopyranose, Mucin-1, ... | | Authors: | Bermejo, I.A, Usabiaga, I, Companon, I, Castro-Lopez, J, Insausti, A, Fernandez, J.A, Avenoza, A, Busto, J.H, Jimenez-Barbero, J, Asensio, J.L, Jimenez-Oses, G, Peregrina, J.M, Hurtado-Guerrero, R, Cocinero, E.J, Corzana, F. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water Sculpts the Distinctive Shapes and Dynamics of the Tumor-Associated Carbohydrate Tn Antigens: Implications for Their Molecular Recognition.
J.Am.Chem.Soc., 140, 2018
|
|
