4MR5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with a quinazolinone ligand (RVX-OH) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 2, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MRD
 
 | | Crystal structure of the murine cd44 hyaluronan binding domain complex with a small molecule | | Descriptor: | CD44 antigen, SULFATE ION, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liu, L.K, Finzel, B. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Identification of an Inducible Binding Site on Cell Surface Receptor CD44 for the Design of Protein-Carbohydrate Interaction Inhibitors.
J.Med.Chem., 57, 2014
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4KW9
 
 | |
4L4S
 
 | | Structural characterisation of the NADH binary complex of human lactate dehydrogenase M isozyme | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LC2
 
 | | Crystal structure of the bromodomain of human BRPF1B | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Peregrin | | Authors: | Tallant, C, Nunez-Alonso, G, Savitsky, P, Picaud, S, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1B
TO BE PUBLISHED
|
|
4L9S
 
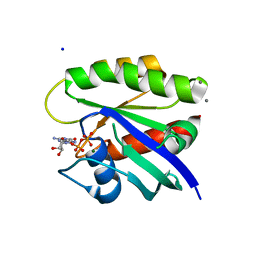 | | Crystal Structure of H-Ras G12C, GDP-bound | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LCH
 
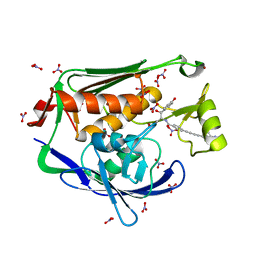 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LKT
 
 | |
4L0E
 
 | |
4KW4
 
 | |
4NP2
 
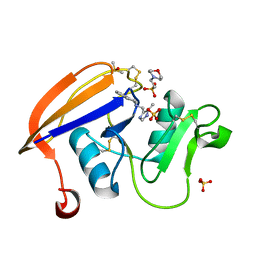 | | Crystal structure of the murine CD44 hyaluronan binding domain complex with a small molecule | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(4-methyl-1H-imidazol-5-yl)methyl]-1,2,3,4-tetrahydroisoquinoline, CD44 antigen, ... | | Authors: | Liu, L.K, Finzel, B. | | Deposit date: | 2013-11-20 | | Release date: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Identification of an Inducible Binding Site on Cell Surface Receptor CD44 for the Design of Protein-Carbohydrate Interaction Inhibitors.
J.Med.Chem., 57, 2014
|
|
4NNI
 
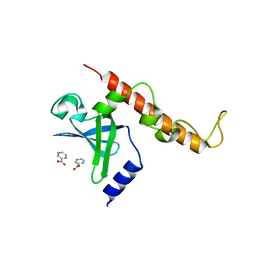 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
4MEQ
 
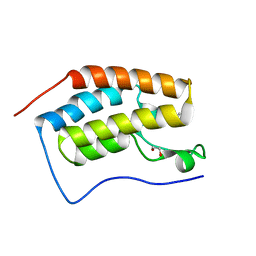 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-7-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4LAA
 
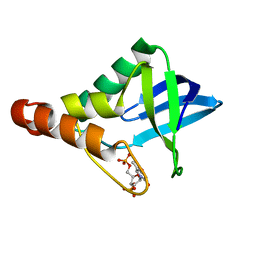 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36H at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Wheeler, E.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B, Robinson, A.C. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36H at cryogenic temperature
To be Published
|
|
4N7Y
 
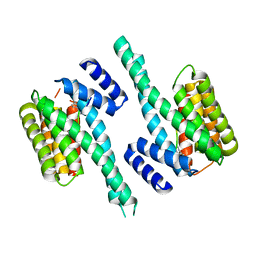 | | Crystal structure of 14-3-3zeta in complex with a 8-carbon-linker cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-16 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4N3N
 
 | |
4NCN
 
 | |
4NCL
 
 | |
4NKL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A/I92Q at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-12 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4NIO
 
 | |
4NMZ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23Q/V66A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cavities in proteins
To be Published
|
|
4NQN
 
 | | Crystal Structure of the bromodomain of human BRD9 in complex with a triazolo-phthalazine ligand | | Descriptor: | 2-chloro-N-[5-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-2-(morpholin-4-yl)phenyl]benzenesulfonamide, Bromodomain-containing protein 9 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-25 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the bromodomain of human BRD9 in complex with a triazolo-phthalazine ligand
To be Published
|
|
4MEO
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4MEP
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3-chloro-pyridone ligand | | Descriptor: | 3-chloro-5-[1-(3-methylpyridin-2-yl)-3-phenyl-1H-1,2,4-triazol-5-yl]pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
