5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
4LWT
 
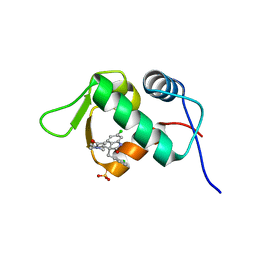 | | The 1.6A Crystal Structure of Humanized Xenopus MDM2 with RO5027344 | | Descriptor: | (3S)-3-[(3R)-1-acetylpiperidin-3-yl]-6-chloro-3-(3-chlorobenzyl)-1,3-dihydro-2H-indol-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Kammlott, U. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
5VO1
 
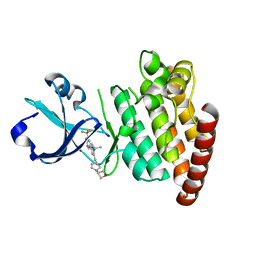 | |
7MAU
 
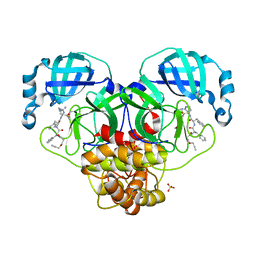 | |
5IS4
 
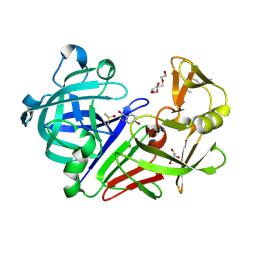 | |
4M8K
 
 | |
4DMZ
 
 | | PelD 156-455 from Pseudomonas aeruginosa PA14, apo form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
2Y2P
 
 | | Penicillin-binding protein 1b (pbp-1b) in complex with an alkyl boronate (z10) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2BHH
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 4-HYDROXYPIPERINDINESULFONYL-INDIRUBINE | | Descriptor: | (2E,3S)-3-HYDROXY-5'-[(4-HYDROXYPIPERIDIN-1-YL)SULFONYL]-3-METHYL-1,3-DIHYDRO-2,3'-BIINDOL-2'(1'H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
8W10
 
 | | Plasmodium vivax PMX-MK7602 inhibitor complex | | Descriptor: | (2R,3S)-3-[(2S)-2-amino-4,4-diethyl-6-oxo-1,3-diazinan-1-yl]-N-[(1R,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-2-(methoxymethyl)-2-methyl-2,3-dihydro-1-benzofuran-5-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2024-02-14 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmodium vivax PMX-MK7602 inhibitor complex
To Be Published
|
|
2BT4
 
 | | Type II Dehydroquinase inhibitor complex | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Toscano, M.D, Stewart, K.A, Coggins, J.R, Lapthorn, A.J, Abell, C. | | Deposit date: | 2005-05-26 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design of New Bifunctional Inhibitors of Type II Dehydroquinase.
Org.Biomol.Chem., 3, 2005
|
|
4LQZ
 
 | |
4MDO
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
8VEB
 
 | |
8VEF
 
 | |
7B6N
 
 | | Crystal structure of MurE from E.coli in complex with Z757284952 | | Descriptor: | 1,2-ETHANEDIOL, N-(2-(2,4-dioxothiazolidin-3-yl)ethyl)-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
2C2M
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-[4-(BENZYLOXY)-4-OXOBUTANOYL]-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14 -PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
4MGV
 
 | |
3T0V
 
 | | Unliganded fluorogen activating protein M8VL | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
9D78
 
 | | Apo-OXA-58 carbapenemase | | Descriptor: | Beta-lactamase | | Authors: | Maggiolo, A.O, Smith, C.A, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
2C2K
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A.J, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2C1E
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Grutter, M.G. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
7MM1
 
 | | PTP1B in complex with TCS401 by Native S-SAD at Room Temperature | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Dalton, K.M, Hekstra, D.R. | | Deposit date: | 2021-04-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Native SAD phasing at room temperature.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7MD7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with triphenylphosphonium analog of chloramphenicol CAM-C4-TPP and protein Y (YfiA) at 2.80A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Pavlova, J.A, Lukianov, D.A, Tereshchenkov, A.G, Makarov, G.I, Khairullina, Z.Z, Tashlitsky, V.N, Paleskava, A, Konevega, A.L, Bogdanov, A.A, Osterman, I.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2021-04-03 | | Release date: | 2021-04-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Triphenylphosphonium Analog of Chloramphenicol upon the Bacterial Ribosome.
Antibiotics, 10, 2021
|
|
