4DQ9
 
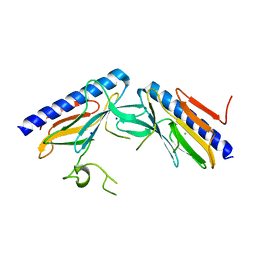 | | Crystal structure of the minor pseudopilin EPSH from the type II secretion system of Vibrio cholerae | | Descriptor: | CHLORIDE ION, General secretion pathway protein H, SODIUM ION | | Authors: | Raghunathan, K, Vago, F.S, Grindem, D, Ball, T, Wedemeyer, W.J, Arvidson, D.N. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The 1.59 angstrom resolution structure of the minor pseudopilin EpsH of Vibrio cholerae reveals a long flexible loop.
Biochim.Biophys.Acta, 1844, 2013
|
|
5I2L
 
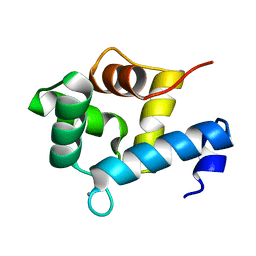 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
5VIX
 
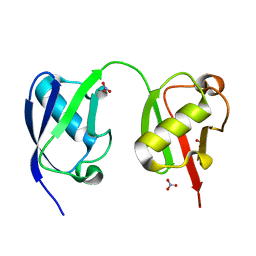 | |
8VCO
 
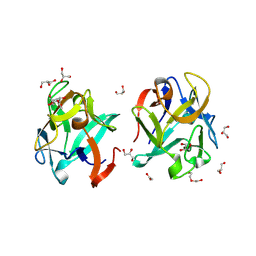 | | Crystal structure of rMcL-1 in complex with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hernandez-Santoyo, A, Loera-Rubalcava, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A mytilectin from Mytilus californianus: Study of its unique galactoside interactions, oligomerization patterns, and antifungal activity.
Int.J.Biol.Macromol., 308, 2025
|
|
4DNF
 
 | |
8VCS
 
 | | Crystal structure of the oligomeric rMcL-1 in complex with lactose | | Descriptor: | CALCIUM ION, Galactose-binding lectin, beta-D-galactopyranose, ... | | Authors: | Hernandez-Santoyo, A, Loera-Rubalcava, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A mytilectin from Mytilus californianus: Study of its unique galactoside interactions, oligomerization patterns, and antifungal activity.
Int.J.Biol.Macromol., 308, 2025
|
|
4M9D
 
 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
5IF7
 
 | |
5VVE
 
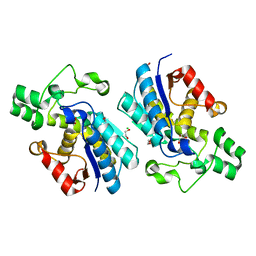 | |
7M49
 
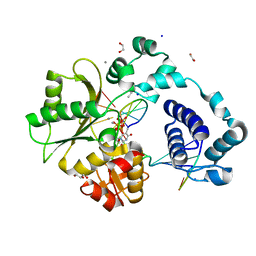 | | DNA Polymerase Lambda, TTP:At Mn2+ Reaction State Ternary Complex, 5 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
8VEZ
 
 | | De novo design apixaban-binding protein: apx1049 | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, De novo design apixaban-binding protein: apx1049 | | Authors: | Bera, A.K, Lee, G.R, Baker, D. | | Deposit date: | 2023-12-20 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Small-molecule binding and sensing with a designed protein family
To Be Published
|
|
5IIS
 
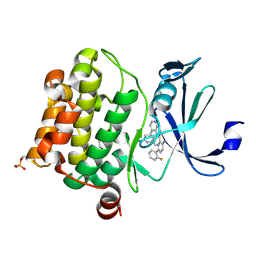 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7M43
 
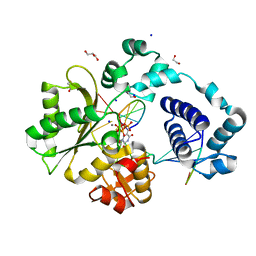 | | DNA Polymerase Lambda, TTP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
2XVD
 
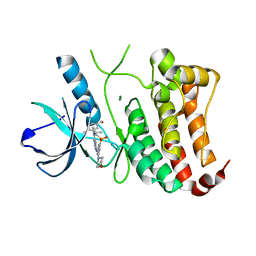 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, {4-METHYL-3-[(1-METHYLETHYL)(2-{[3-(METHYLSULFONYL)-5-MORPHOLIN-4-YLPHENYL]AMINO}PYRIMIDIN-4-YL)AMINO]PHENYL}METHANOL | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L. | | Deposit date: | 2010-10-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 4: Discovery and Optimization of a Benzylic Alcohol Series.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2BQW
 
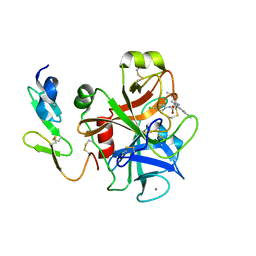 | | CRYSTAL STRUCTURE OF FACTOR XA IN COMPLEX WITH COMPOUND 45 | | Descriptor: | 1-{2-[(4-CHLOROPHENYL)AMINO]-2-OXOETHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-28 | | Release date: | 2006-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
7M4K
 
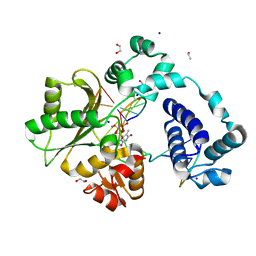 | | DNA Polymerase Lambda, TTPaS:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
2XZZ
 
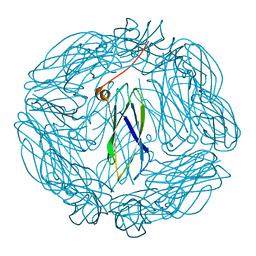 | | Crystal structure of the human transglutaminase 1 beta-barrel domain | | Descriptor: | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE K | | Authors: | Vollmar, M, Krysztofinska, E, Krojer, T, Yue, W.W, Cooper, C, Kavanagh, K, Allerston, C, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Human Transglutaminase 1 Beta-Barrel Domain
To be Published
|
|
7M4L
 
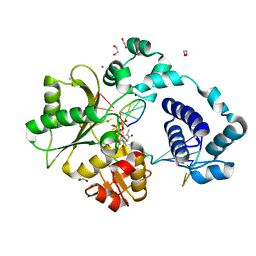 | | DNA Polymerase Lambda, TTPaS:At Mn2+ Product State Ternary Complex, 60 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*(YQS))-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
4DEH
 
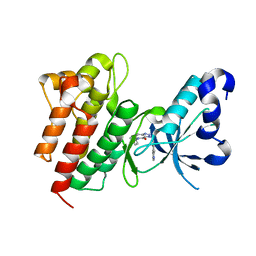 | | Crystal structure of c-Met in complex with triazolopyridinone inhibitor 3 | | Descriptor: | 5-phenyl-3-(quinolin-6-ylmethyl)-3,5,6,7-tetrahydro-4H-[1,2,3]triazolo[4,5-c]pyridin-4-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Bellon, S.F, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2XIJ
 
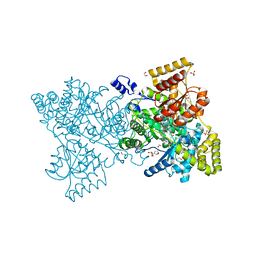 | | Crystal structure of human methylmalonyl-CoA mutase in complex with adenosylcobalamin | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXYADENOSINE, ... | | Authors: | Yue, W.W, Froese, D.S, Kochan, G, Chaikuad, A, Krojer, T, Muniz, J, Vollmar, M, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Human Gtpase Mmaa and Vitamin B12-Dependent Methylmalonyl-Coa Mutase and Insight Into Their Complex Formation.
J.Biol.Chem., 285, 2010
|
|
7M6Q
 
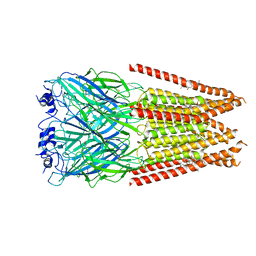 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
5IQ6
 
 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | Authors: | Tarantino, D, Mastrangelo, E, Milani, M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
4DI0
 
 | |
7LXR
 
 | |
