7WUC
 
 | |
7X5T
 
 | |
7WWS
 
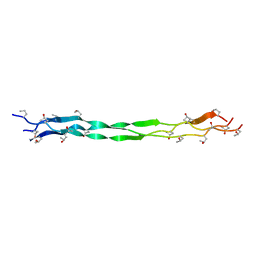 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
5VA0
 
 | |
7XF7
 
 | |
7XF6
 
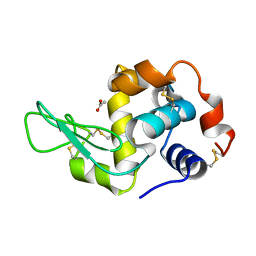 | | Crystal Structure of Human Lysozyme | | Descriptor: | ACETATE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Human Lysozyme Complexed with N-Acetyl-alpha-d-glucosamine.
Appl Sci (Basel), 12, 2022
|
|
7WVM
 
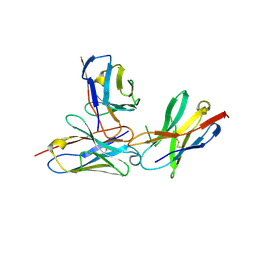 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
7WZE
 
 | |
7XF8
 
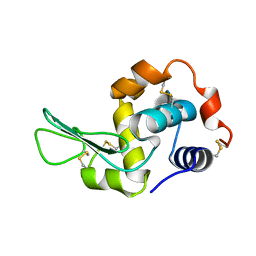 | |
7KJZ
 
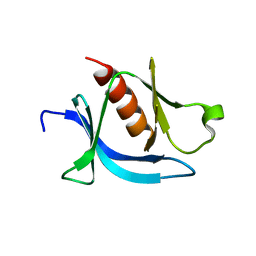 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7X2U
 
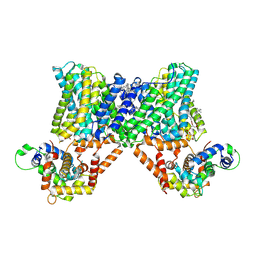 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | Authors: | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-02-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
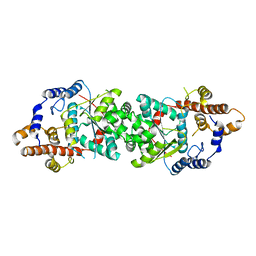
5UJI
 
 | |
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-04-14 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L5S
 
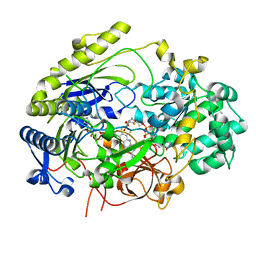 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
5V35
 
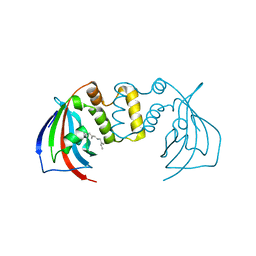 | | Crystal structure of V71F mutant of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7X4O
 
 | |
5V75
 
 | |
5UK6
 
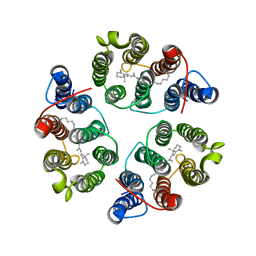 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy and DEER | | Descriptor: | Bacteriorhodopsin | | Authors: | Milikisiyants, S, Wang, S, Munro, R.A, Donohue, M, Ward, M.E, Brown, L.S, Smirnova, T.I, Ladizhansky, V, Smirnov, A.I. | | Deposit date: | 2017-01-20 | | Release date: | 2017-05-31 | | Last modified: | 2020-01-08 | | Method: | SOLID-STATE NMR | | Cite: | Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints.
J. Mol. Biol., 429, 2017
|
|
5UOP
 
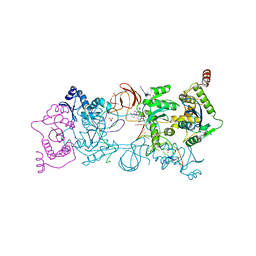 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 18) | | Descriptor: | (1S,2S,5R)-8'-[(3-chloro-4-fluorophenyl)methyl]-2'-[2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl]-6'-hydroxy-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7L7C
 
 | |
5VC4
 
 | | Crystal structure of HUMAN WEE1 KINASE domain in complex with Bosutinib-isomer | | Descriptor: | 4-[(3,5-DICHLORO-4-METHOXYPHENYL)AMINO]-6-METHOXY-7-[3-(4-METHYLPIPERAZIN-1-YL)PROPOXY]QUINOLINE-3-CARBONITRILE, PHOSPHATE ION, Wee1-like protein kinase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
7L76
 
 | |
5VJU
 
 | | De Novo Photosynthetic Reaction Center Protein Variant Equipped with His-Tyr H-bond, Heme B, and Cd(II) ions | | Descriptor: | CADMIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette Leu71His variant | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VPG
 
 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
