2ICX
 
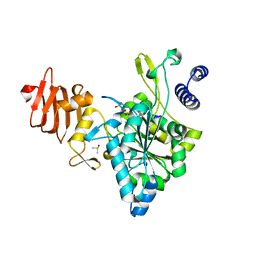 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UTP | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE 5'-TRIPHOSPHATE | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
2ICY
 
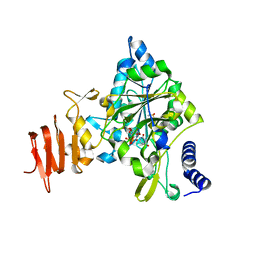 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UDP-glucose | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
2ICZ
 
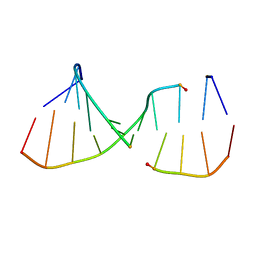 | | NMR Structures of the Expanded DNA 10bp xTGxTAxCxGCxAxGT:xACTxGCGxTAxCA | | Descriptor: | 5'-D(*(XAE)P*CP*TP*(XGA)P*CP*GP*(XTY)P*AP*(XCS)P*A)-3', 5'-D(*(XTY)P*GP*(XTY)P*AP*(XCS)P*(XGA)P*CP*(XAE)P*(XGA)P*T)-3' | | Authors: | Lynch, S.R. | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Toward a Designed, Functioning Genetic System with Expanded-Size Base Pairs: Solution Structure of the Eight-Base xDNA Double Helix.
J.Am.Chem.Soc., 128, 2006
|
|
2ID0
 
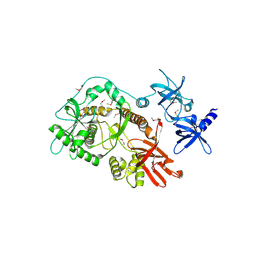 | | Escherichia coli RNase II | | Descriptor: | Exoribonuclease 2, MANGANESE (II) ION | | Authors: | Zuo, Y, Zhang, J, Wang, Y, Malhotra, A. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Processivity and Single-Strand Specificity of RNase II.
Mol.Cell, 24, 2006
|
|
2ID1
 
 | | X-Ray Crystal Structure of Protein CV0518 from Chromobacterium violaceum, Northeast Structural Genomics Consortium Target CvR5. | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Forouhar, F, Zhou, W, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: |
|
|
2ID2
 
 | | GAPN T244S mutant X-ray structure at 2.5 A | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pailot, A, D'Ambrosio, K, Corbier, C, Talfournier, F, Branlant, G. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Invariant Thr(244) is essential for the efficient acylation step of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans.
Biochem.J., 400, 2006
|
|
2ID3
 
 | | Crystal structure of transcriptional regulator SCO5951 from Streptomyces coelicolor A3(2) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative transcriptional regulator | | Authors: | Grabowski, M, Chruszcz, M, Koclega, K.D, Cymborowski, M, Gu, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: |
|
|
2ID4
 
 | |
2ID5
 
 | | Crystal Structure of the Lingo-1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | Authors: | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | Deposit date: | 2006-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
2ID6
 
 | |
2ID7
 
 | | 1.75 A Structure of T87I Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID8
 
 | | Crystal structure of Proteinase K | | Descriptor: | (S)-(2,3-DIHYDROXYPROPOXY)TRIHYDROXYBORATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, J, Dauter, M, Dauter, Z. | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | What can be done with a good crystal and an accurate beamline?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2ID9
 
 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2IDA
 
 | | Solution NMR Structure of Protein RPA1320 from Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RpT3; Ontario Center for Structural Proteomics Target RP1313. | | Descriptor: | Hypothetical protein, ZINC ION | | Authors: | Lemak, A, Yee, A, Lukin, J.A, Karra, M, Gutmanas, A, Guido, V, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RPA1320
To be Published
|
|
2IDB
 
 | | Crystal Structure of 3-octaprenyl-4-hydroxybenzoate decarboxylase (UbiD) from Escherichia coli, Northeast Structural Genomics Target ER459. | | Descriptor: | 1,2-ETHANEDIOL, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Chen, C.X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of 3-octaprenyl-4-hydroxybenzoate decarboxylase (UbiD) from Escherichia coli, Northeast Structural Genomics Target ER459.
TO BE PUBLISHED
|
|
2IDC
 
 | | Structure of the Histone H3-Asf1 Chaperone Interaction | | Descriptor: | ANTI-SILENCING PROTEIN 1 AND HISTONE H3 CHIMERA | | Authors: | Antczak, A.J, Tsubota, T, Kaufman, P.D, Berger, J.M. | | Deposit date: | 2006-09-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics.
Bmc Struct.Biol., 6, 2006
|
|
2IDE
 
 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
2IDF
 
 | | P. aeruginosa azurin N42C/M64E double mutant, BMME-linked dimer | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | Einsle, O, de Jongh, T.E, Hoffmann, M, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-09-15 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electron transfer in a crosslinked protein dimer mediated by a hydrogen-bonded network across the dimer interface
To be Published
|
|
2IDG
 
 | | Crystal Structure of hypothetical protein AF0160 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0160 | | Authors: | Zhao, M, Zhang, M, Chang, J, Chen, L, Xu, H, Li, Y, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Hypothetical Protein AF0160 from Archaeoglobus fulgidus at 2.69 Angstrom resolution
To be Published
|
|
2IDH
 
 | |
2IDJ
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Apoprotein, Monoclinic Form | | Descriptor: | CALCIUM ION, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
2IDK
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Complexed With Folate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
2IDL
 
 | | Crystal Structure of Conserved Protein of Unknown Function from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, Hypothetical protein, SODIUM ION | | Authors: | Nocek, B, Wu, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2IDN
 
 | | NMR structure of a new modified Thrombin Binding Aptamer containing a 5'-5' inversion of polarity site | | Descriptor: | 3'-D(P*GP*G*T)-5'-5'-D(P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3' | | Authors: | Randazzo, A, Martino, L, Virno, A, Mayol, L, Giancola, C. | | Deposit date: | 2006-09-15 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site.
Nucleic Acids Res., 34, 2006
|
|
