5U0J
 
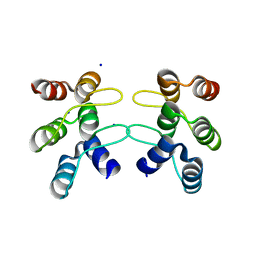 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - monoclinic crystal form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SODIUM ION | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5U0K
 
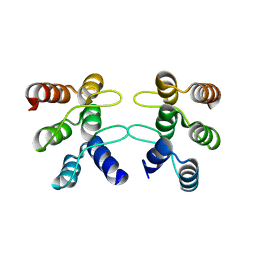 | | C-terminal ankyrin repeats from human liver-type glutaminase (GAB/LGA) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Ferreira, I.M, Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5AO6
 
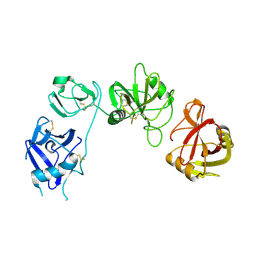 | | Endo180 D1-4, trigonal form | | Descriptor: | C-TYPE MANNOSE RECEPTOR 2 | | Authors: | Paracuellos, P, Briggs, D.C, Carafoli, F, Loncar, T, Hohenester, E. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Insights Into Collagen Uptake by C-Type Mannose Receptors from the Crystal Structure of Endo180 Domains 1-4.
Structure, 23, 2015
|
|
5AO5
 
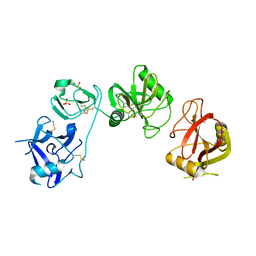 | | Endo180 D1-4, monoclinic form | | Descriptor: | C-TYPE MANNOSE RECEPTOR 2, SODIUM ION, SULFATE ION | | Authors: | Paracuellos, P, Briggs, D.C, Carafoli, F, Loncar, T, Hohenester, E. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Insights Into Collagen Uptake by C-Type Mannose Receptors from the Crystal Structure of Endo180 Domains 1-4.
Structure, 23, 2015
|
|
5NB3
 
 | | High resolution C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
2RFS
 
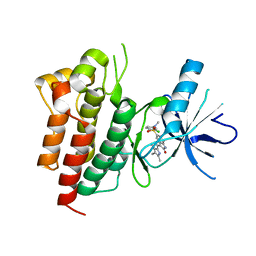 | | X-ray structure of SU11274 bound to c-Met | | Descriptor: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
2RFN
 
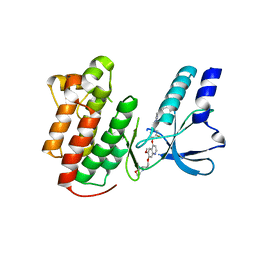 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
2KQF
 
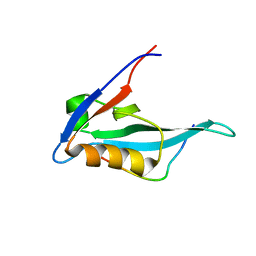 | | Solution structure of MAST205-PDZ complexed with the C-terminus of a rabies virus G protein | | Descriptor: | C-terminal motif from Glycoprotein, Microtubule-associated serine/threonine-protein kinase 2 | | Authors: | Terrien, E, Wolff, N, Cordier, F, Simenel, C, Bernard, A, Lafon, M, Delepierre, M. | | Deposit date: | 2009-11-04 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the PDZ of Microtubule-associated serine/threonine kinase 205 (MAST205) in complex with the C-Terminal motif from the rabies virus glycoprotein
To be Published
|
|
2KYL
 
 | | Solution structure of MAST2-PDZ complexed with the C-terminus of PTEN | | Descriptor: | C-terminus of Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, Microtubule-associated serine/threonine-protein kinase 2 | | Authors: | Terrien, E, Wolff, N, Cordier, F, Simenel, C, Lafon, M, Delepierre, M. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MAST2-PDZ complexed with the C-terminus of PTEN
To be Published
|
|
1A1A
 
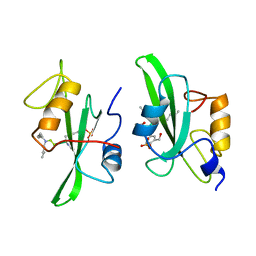 | |
1ZMF
 
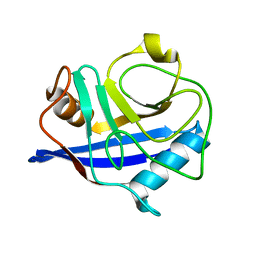 | | C domain of human cyclophilin-33(hcyp33) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Wang, T, Yun, C.-H, Gu, S.-Y, Chang, W.-R, Liang, D.-C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88A crystal structure of the C domain of hCyP33: A novel domain of peptidyl-prolyl cis-trans isomerase
Biochem.Biophys.Res.Commun., 333, 2005
|
|
4FJ3
 
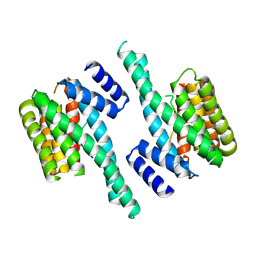 | |
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
2LS8
 
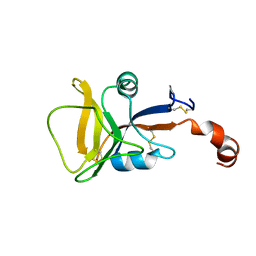 | | Solution structure of human C-type lectin domain family 4 member D | | Descriptor: | C-type lectin domain family 4 member D | | Authors: | Harris, R, Gaudette, J, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human C-type lectin domain family 4 member D
To be Published
|
|
1A08
 
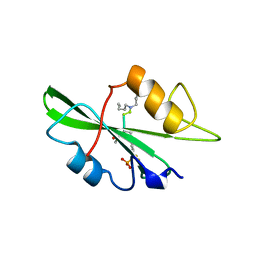 | |
1A1B
 
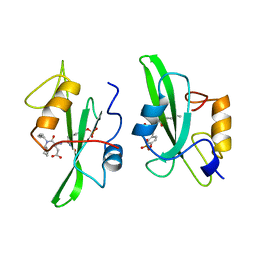 | |
1A07
 
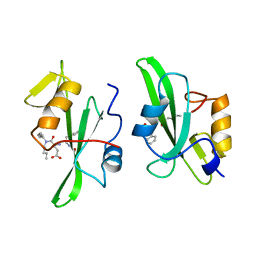 | |
1A1C
 
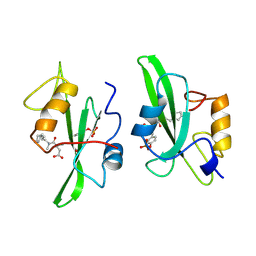 | |
1HZL
 
 | | SOLUTION STRUCTURES OF C-1027 APOPROTEIN AND ITS COMPLEX WITH THE AROMATIZED CHROMOPHORE | | Descriptor: | C-1027 APOPROTEIN, C-1027 AROMATIZED CHROMOPHORE | | Authors: | Tanaka, T, Fukuda-Ishisaka, S, Hirama, M, Otani, T. | | Deposit date: | 2001-01-25 | | Release date: | 2001-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structures of C-1027 apoprotein and its complex with the aromatized chromophore.
J.Mol.Biol., 309, 2001
|
|
1YUA
 
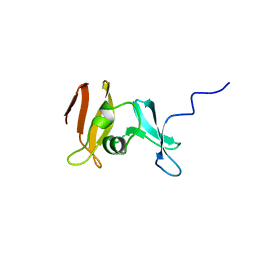 | |
1A09
 
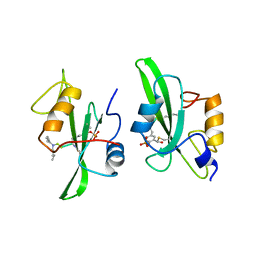 | |
1A1E
 
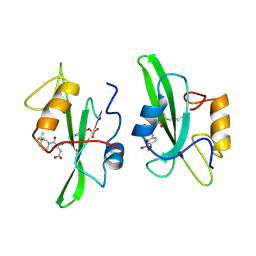 | |
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | Descriptor: | ACETATE ION, Oxidoreductase, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
2YY0
 
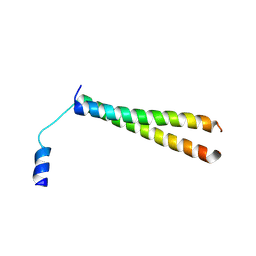 | | Crystal Structure of MS0802, c-Myc-1 binding protein domain from Homo sapiens | | Descriptor: | C-Myc-binding protein | | Authors: | Xie, Y, Wang, H, Ihsanawati, K.T, Kishishita, S, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | crystal structure of c-Myc-1 binding protein domain from Homo sapiens
To be Published
|
|
1CMX
 
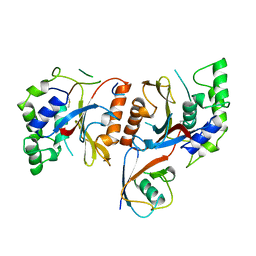 | |
