8GXK
 
 | | Pseudomonas jinjuensis N-acetyltransferase | | Descriptor: | COENZYME A, Protein N-acetyltransferase, RimJ/RimL family | | Authors: | Song, Y.J, Bao, R. | | Deposit date: | 2022-09-20 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur.
Nucleic Acids Res., 50, 2022
|
|
8GXF
 
 | | Pseudomonas flexibilis GCN5 family acetyltransferase | | Descriptor: | COENZYME A, GCN5 family acetyltransferase | | Authors: | Song, Y.J, Bao, R. | | Deposit date: | 2022-09-20 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur.
Nucleic Acids Res., 50, 2022
|
|
8GXJ
 
 | |
6YCA
 
 | | Crystal structure of Eis1 from Mycobacterium abscessus | | Descriptor: | ACETYL COENZYME *A, SULFATE ION, Uncharacterized N-acetyltransferase D2E76_00625 | | Authors: | Blaise, M, Ung, K.L. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the N-acetyltransferase Eis1 from Mycobacterium abscessus reveals the molecular determinants of its incapacity to modify aminoglycosides.
Proteins, 89, 2021
|
|
2HEO
 
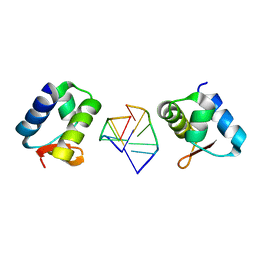 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | Authors: | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
6QYA
 
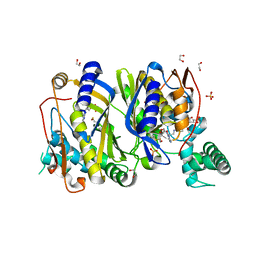 | |
5AHU
 
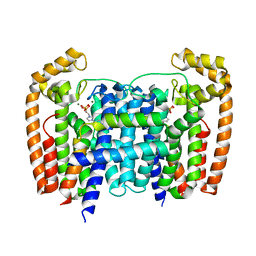 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-1326 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PUTATIVE, MAGNESIUM ION, ... | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2015-02-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
5AB5
 
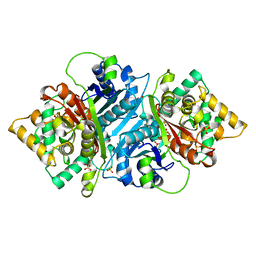 | |
7S6C
 
 | | CryoEM structure of modular PKS holo-Lsd14 stalled at the condensation step and bound to antibody fragment 1B2, composite structure | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 4'-PHOSPHOPANTETHEINE, 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Bagde, S.R, Kim, C.-Y, Fromme, J.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modular polyketide synthase contains two reaction chambers that operate asynchronously.
Science, 374, 2021
|
|
6SWK
 
 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus | | Descriptor: | Histidine kinase | | Authors: | Lansky, S, Shiradsky, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus
To Be Published
|
|
7S6D
 
 | | CryoEM structure of modular PKS holo-Lsd14 bound to antibody fragment 1B2, composite structure | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-deoxyerythronolide-B synthase EryA2, modules 3 and 4, ... | | Authors: | Bagde, S.R, Kim, C.-Y, Fromme, J.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modular polyketide synthase contains two reaction chambers that operate asynchronously.
Science, 374, 2021
|
|
2HG9
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with tetrabrominated phosphatidylcholine | | Descriptor: | (7R,18S,19R)-18,19-DIBROMO-7-{[(9S,10S)-9,10-DIBROMOOCTADECANOYL]OXY}-4-HYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-P HOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
7RUV
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase E193K bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
6SMO
 
 | | AntDE:AntF (apo): type II PKS acyl-carrier protein in complex with its ketosynthase bound to the hexaketide | | Descriptor: | 3,5,7,9,11-pentakis(oxidanylidene)dodecanal, Acyl carrier protein, Ketoacyl_synth_N domain-containing protein, ... | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
7RUU
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7SA3
 
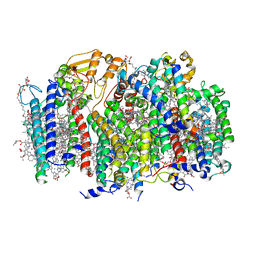 | | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
J.Biol.Chem., 298, 2021
|
|
2HHK
 
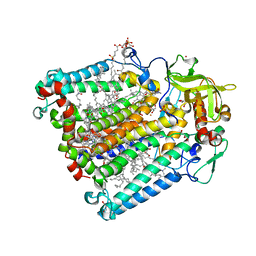 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylglycerol | | Descriptor: | (1R)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
7SGY
 
 | |
8CU9
 
 | |
6RXT
 
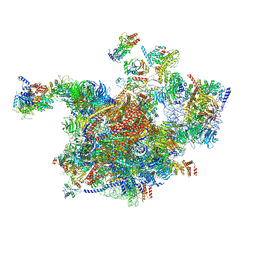 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
2HG3
 
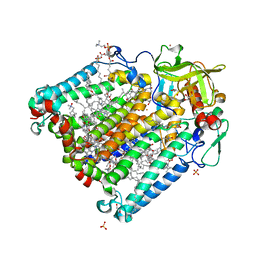 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with brominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
1A2N
 
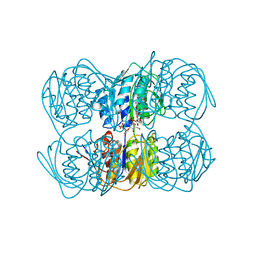 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
5D84
 
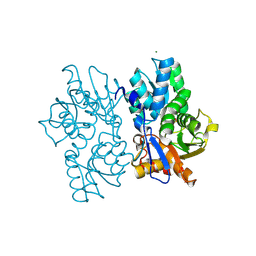 | | Staphyloferrin B precursor biosynthetic enzyme SbnA bound to PLP | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Probable siderophore biosynthesis protein SbnA | | Authors: | Grigg, J.C, Kobylarz, M.J, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
6RXZ
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
