6N4Q
 
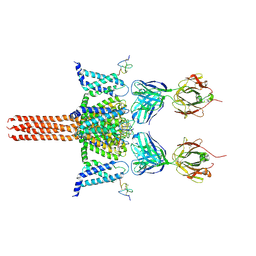 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
8GJ8
 
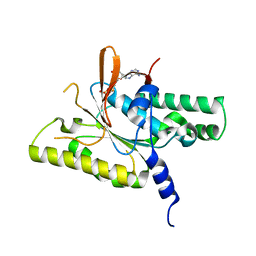 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
6PX4
 
 | |
8FZA
 
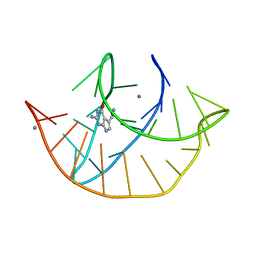 | | Class I type III preQ1 riboswitch from E. coli | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PreQ1 Riboswitch (30-MER) | | Authors: | Wedekind, J.E, Schroeder, G.M, Jenkins, J.L. | | Deposit date: | 2023-01-27 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function analysis of a type III preQ 1 -I riboswitch from Escherichia coli reveals direct metabolite sensing by the Shine-Dalgarno sequence.
J.Biol.Chem., 299, 2023
|
|
6N8I
 
 | |
7BBC
 
 | | Joint X-ray/neutron room temperature structure of perdeuterated PLL lectin in complex with perdeuterated L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (1.84 Å), X-RAY DIFFRACTION | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6X3L
 
 | | Sortilin-Progranulin Interaction With Compound 2 | | Descriptor: | 1-benzyl-3-tert-butyl-1H-pyrazole-5-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8GTG
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
6MT4
 
 | |
6Q4E
 
 | | CDK2 in complex with FragLite33 | | Descriptor: | 6-iodanyl-7~{H}-purin-2-amine, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4K
 
 | | CDK2 in complex with FragLite38 | | Descriptor: | (~{E})-3-[3-[(4-chlorophenyl)carbamoyl]phenyl]prop-2-enoic acid, 1,2-ETHANEDIOL, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6MS9
 
 | | GDP-bound KRAS P34R mutant | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Bera, A.K, Westover, K.D. | | Deposit date: | 2018-10-16 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | GTP hydrolysis is modulated by Arg34 in the RASopathy-associated KRASP34R.
Birth Defects Res, 112, 2020
|
|
8TC5
 
 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
6WG5
 
 | | Human ectonucleoside triphosphate diphosphohydrolase 4 (ENTPD4, NTPDase 4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ectonucleoside triphosphate diphosphohydrolase 4 | | Authors: | Gorelik, A, Labriola, J.M, Illes, K, Nagar, B. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the nucleotide-metabolizing enzyme NTPDase4.
Protein Sci., 29, 2020
|
|
6MUM
 
 | | Murine PI3K delta kinsae domain - cpd 3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, [(3S)-3-{[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]oxy}pyrrolidin-1-yl](oxan-4-yl)methanone | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
8GTI
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GSM
 
 | | Crystal Structure of VibMO1 | | Descriptor: | 4-hydroxybenzoate decarboxylase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ying, Z, Feng, K.N. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal Structure of VibMO1
To Be Published
|
|
8GTM
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
7BIR
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1-[[(1~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-1-(4-chlorophenyl)-7-fluoranyl-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-1-yl]oxymethyl]cyclopropane-1-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6Z1E
 
 | |
6NP6
 
 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NPL
 
 | | Cryo-EM structure of NKCC1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6NPH
 
 | | Structure of NKCC1 TM domain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6YVW
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with monocyclic BB-328 | | Descriptor: | 4-[(5-bromanyl-4,6-dimethyl-pyridin-2-yl)amino]-4-oxidanylidene-butanoic acid, Egl nine homolog 1, FE (III) ION, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
