7TVA
 
 | | Stat5a Core in complex with AK2292 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
8S4A
 
 | | CdaA-B04 complex | | Descriptor: | 2-[cyanomethyl(methyl)amino]-N-(6-methylpyridin-2-yl)ethanamide, CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | CdaA-B04 complex
To Be Published
|
|
8RUN
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Domains 1-5, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
7TVB
 
 | | Stat5A Core in Complex with AK305 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
8S4B
 
 | | CdaA-C08 complex | | Descriptor: | (4-methoxycarbonylphenyl)methylazanium, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CdaA-C08 complex
To Be Published
|
|
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
8S4C
 
 | | CdaA-H04 complex | | Descriptor: | DAMINOZIDE, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CdaA-H04 complex
To Be Published
|
|
8HDD
 
 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Sode, K. | | Deposit date: | 2022-11-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
8S48
 
 | | CdaA-C11 complex | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | CdaA-C11 complex
To Be Published
|
|
7THG
 
 | |
7TYQ
 
 | | TEAD2 bound to Compound 1 | | Descriptor: | Transcriptional enhancer factor TEF-4, ethyl (8S)-7-oxo-5-[4-(trifluoromethyl)phenyl]-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carboxylate | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYU
 
 | | TEAD2 bound to Compound 2 | | Descriptor: | (3R)-1-[(8S)-5-(4-cyclohexylphenyl)-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonyl]pyrrolidine-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYP
 
 | | TEAD2 bound to GNE-7883 | | Descriptor: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7U37
 
 | |
6VY2
 
 | |
8S65
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) as target for anti Toxoplasma gondii compounds: crystal structure, biochemical characterization and biological evaluation of inhibitors | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Mazzone, F, Hoeppner, A, Reiners, J, Applegate, V, Abdullaziz, M, Gottstein, J, Wesemann, M, Kurz, T, Smits, S.H, Pfeffer, K. | | Deposit date: | 2024-02-26 | | Release date: | 2024-08-21 | | Method: | SOLUTION SCATTERING (2.56 Å), X-RAY DIFFRACTION | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) as target for anti Toxoplasma gondii agents: crystal structure, biochemical characterisation and biological evaluation of inhibitors.
Biochem.J., 2024
|
|
6VUI
 
 | | Metabolite-bound PreQ1 riboswitch with Mn2+ | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
8FIE
 
 | |
6VI1
 
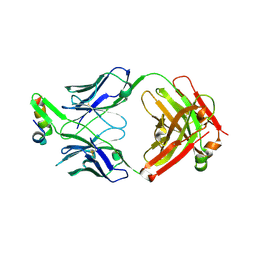 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8T62
 
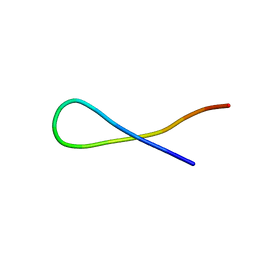 | |
6VTN
 
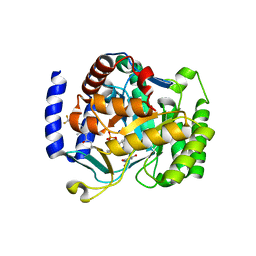 | |
8EQD
 
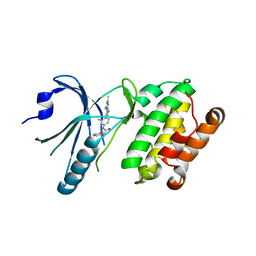 | | Co-crystal structure of PERK with compound 24 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
6VZ9
 
 | |
6W1E
 
 | |
8F2G
 
 | | Crystal structure of Hen Egg White Lysozyme at 0.44 GPa | | Descriptor: | Lysozyme C | | Authors: | Marshall, A.C, Boer, S.A, Turner, G, Moggach, S.A, Bond, C.S, Vrielink, A. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-pressure single-crystal diffraction at the Australian Synchrotron.
J.Synchrotron Radiat., 30, 2023
|
|
