5CNY
 
 | |
6GOF
 
 | | KRAS full length G12D GPPNHP | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Ehebauer, M.T, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3CUL
 
 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (92-MER), ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
6GOH
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
7ZLQ
 
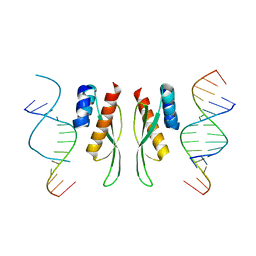 | |
7ZJ1
 
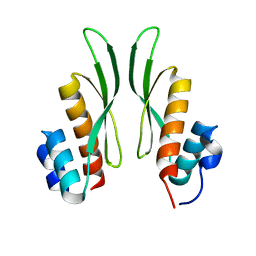 | | Crystal structure of ADAR1-dsRBD3 dimer | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Mboukou, A, Barraud, P. | | Deposit date: | 2022-04-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dimerization of ADAR1 modulates site-specificity of RNA editing
Biorxiv, 2023
|
|
3CV8
 
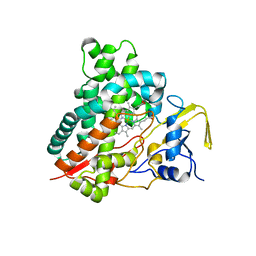 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84F mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
7ZRR
 
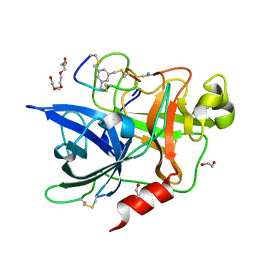 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, AMINO GROUP, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965
To Be Published
|
|
7ZRT
 
 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970
To Be Published
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
6TS4
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
3CWL
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
6GUH
 
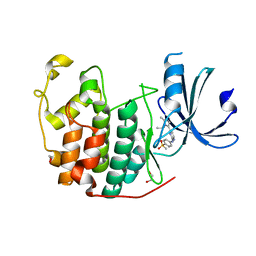 | | CDK2 in complex with AZD5438 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
3CXV
 
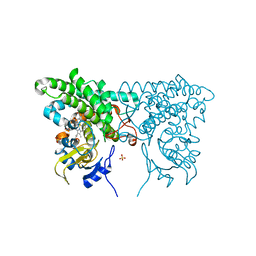 | |
3CY5
 
 | |
3CON
 
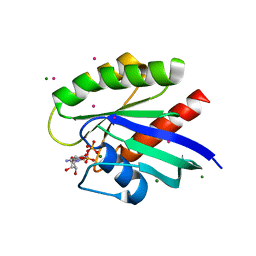 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
3CPP
 
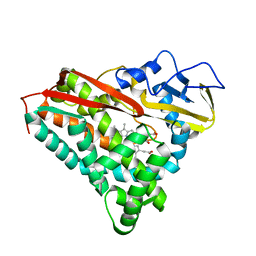 | |
5COO
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
6GJB
 
 | | Erk2 signalling protein | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[4-[1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-2-oxidanylidene-pyridin-4-yl]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Quantitation of ERK1/2 inhibitor cellular target occupancies with a reversible slow off-rate probe.
Chem Sci, 9, 2018
|
|
3D15
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-(3-chloro-phenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)- ethyl]-thiazol-2-yl}-urea [SNS-314] | | Descriptor: | 1-(3-chlorophenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Bui, M, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6TS7
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-(1,2,3,4-tetrahydroisoquinolin-7-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
3CQX
 
 | | Chaperone Complex | | Descriptor: | BAG family molecular chaperone regulator 2, Heat shock cognate 71 kDa protein, SODIUM ION, ... | | Authors: | Xu, Z, Nix, J.C, Misra, S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2
Nat.Struct.Mol.Biol., 15, 2008
|
|
5CE4
 
 | | High Resolution X-Ray and Neutron diffraction structure of H-FABP | | Descriptor: | Fatty acid-binding protein, heart, OLEIC ACID | | Authors: | Podjarny, A.D, Howard, E.I, Blakeley, M.P, Guillot, B. | | Deposit date: | 2015-07-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (0.98 Å), X-RAY DIFFRACTION | | Cite: | High-resolution neutron and X-ray diffraction room-temperature studies of an H-FABP-oleic acid complex: study of the internal water cluster and ligand binding by a transferred multipolar electron-density distribution.
Iucrj, 3, 2016
|
|
3D22
 
 | | Crystal structure of a poplar thioredoxin h mutant, PtTrxh4C61S | | Descriptor: | PHOSPHATE ION, Thioredoxin H-type | | Authors: | Koh, C.S, Didierjean, C, Corbier, C, Rouhier, N, Jacquot, J.P, Gelhaye, E. | | Deposit date: | 2008-05-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Atypical Catalytic Mechanism Involving Three Cysteines of Thioredoxin.
J.Biol.Chem., 283, 2008
|
|
