6M4V
 
 | | Crystal structure of MBP fused split FKBP in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6M10
 
 | | Crystal structure of PA4853 (Fis) from Pseudomonas aeruginosa | | Descriptor: | Putative Fis-like DNA-binding protein | | Authors: | Zhang, H, Gao, Z, Zhou, J, Dong, Y. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of the nucleoid-associated protein Fis (PA4853) from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6LOL
 
 | | The crystal structure of full length IpaH9.8 | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
5C5T
 
 | |
5C6I
 
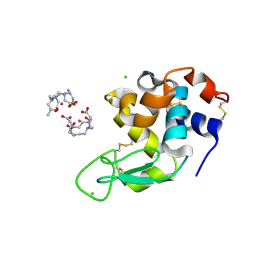 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
8I55
 
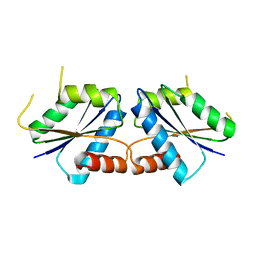 | |
8I56
 
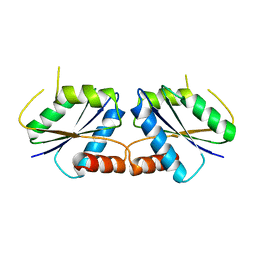 | | CfbA S11T variant | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Ogawa, S, Fujishiro, T. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Substrate selectivity of CfbA
To be published
|
|
8I58
 
 | | Uroporphyrin I (UPI)-bound CfbA | | Descriptor: | 3-[(1Z,9Z)-3,8,13,18-tetrakis(2-hydroxy-2-oxoethyl)-7,12,17-tris(3-hydroxy-3-oxopropyl)-21,23-dihydroporphyrin-2-yl]propanoic acid, Sirohydrochlorin cobaltochelatase | | Authors: | Ogawa, S, Fujishiro, T. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Substrate selectivity of CfbA
To be published
|
|
5BY1
 
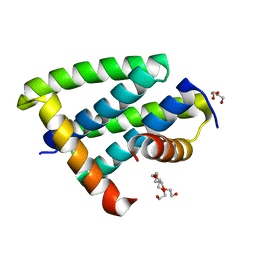 | | H18 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, PENTAETHYLENE GLYCOL | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
6M06
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(Z)-1,3-bis(oxidanyl)-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M25
 
 | | Sirohydrochlorin nickelochelatase CfbA in P41 space group | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M27
 
 | | Sirohydrochlorin nickelochelatase CfbA in complex with Ni2+ | | Descriptor: | NICKEL (II) ION, Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
8I57
 
 | | Uroporphyrin III (UPIII)-bound CfbA | | Descriptor: | 3,3',3'',3'''-[3,8,13,17-tetrakis(carboxymethyl)porphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Sirohydrochlorin cobaltochelatase | | Authors: | Ogawa, S, Fujishiro, T. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Substrate selectivity of CfbA
To be published
|
|
7PP0
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 28 (JMV-7038) | | Descriptor: | 2-[2-[3-[3-(2-morpholin-4-ylethoxy)phenyl]-5-sulfanylidene-1H-1,2,4-triazol-4-yl]ethyl]benzoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Chelini, G, De Luca, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-beta-Lactamase Inhibitors.
Chemmedchem, 17, 2022
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6MBT
 
 | | Crystal structure of wild-type KRAS bound to GDP and Mg (Space group C2) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
5CCI
 
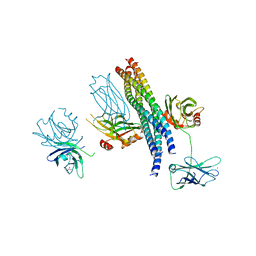 | | Structure of the Mg2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | MAGNESIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
8I9B
 
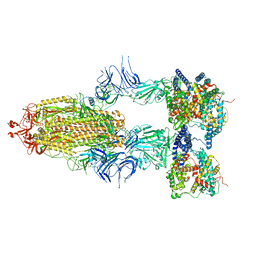 | | S-ECD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
5BW2
 
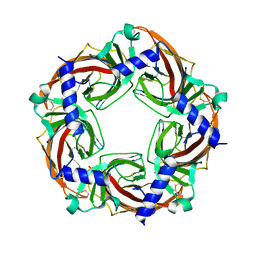 | | X-ray crystal structure of Aplysia californica acetylcholine binding protein (Ac-AChBP) Y55W in complex with 2-Pyridin-3-yl-1-aza-bicyclo[2.2.2]octane; 2-(3-pyridyl)quinuclidine; 2-PQ (TI-4699) | | Descriptor: | (2R)-2-(pyridin-3-yl)-1-azabicyclo[2.2.2]octane, Soluble acetylcholine receptor | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-05 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
6M18
 
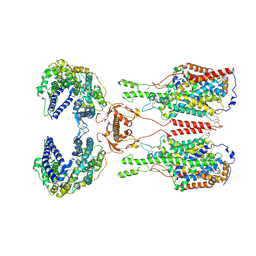 | | ACE2-B0AT1 complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
6M2A
 
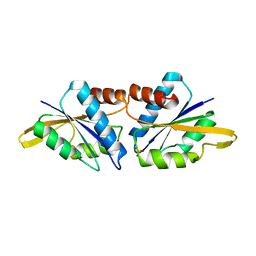 | |
5C6B
 
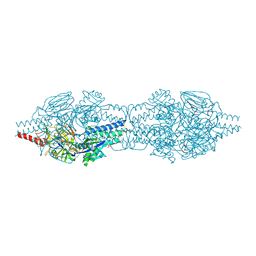 | |
6M1D
 
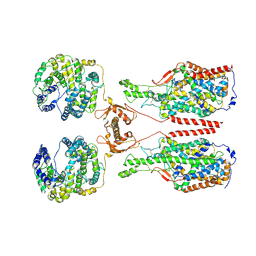 | | ACE2-B0AT1 complex, open conformation | | Descriptor: | Angiotensin-converting enzyme 2, Sodium-dependent neutral amino acid transporter B(0)AT1 | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
7PWY
 
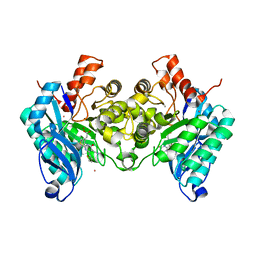 | | Structure of human dimeric ACMSD in complex with the inhibitor TES-1025 | | Descriptor: | 2-[3-[(5-cyano-6-oxidanylidene-4-thiophen-2-yl-1H-pyrimidin-2-yl)sulfanylmethyl]phenyl]ethanoic acid, 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, POTASSIUM ION, ... | | Authors: | Cianci, M, Giacche, N, Carotti, A, Liscio, P, Amici, A, Cialabrini, L, De Franco, F, Pellicciari, R, Raffaelli, N. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Human Dimeric alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Inhibition With TES-1025.
Front Mol Biosci, 9, 2022
|
|
6M7H
 
 | | Structure of calmodulin with KN93 | | Descriptor: | CALCIUM ION, Calmodulin-1, N-[2-[[[3-(4'-Chlorophenyl)-2-propenyl]methylamino]methyl]phenyl]-N-(2-hydroxyethyl)-4'-methoxybenzenesulfonamide | | Authors: | Damo, S.M, Pattanayek, R, Johnson, C.N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1.5 and RyR2 function.
Cell Calcium, 82, 2019
|
|
