1VFM
 
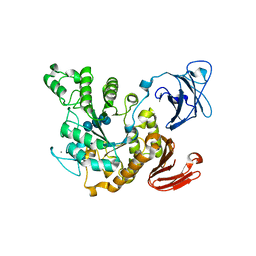 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/alpha-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFU
 
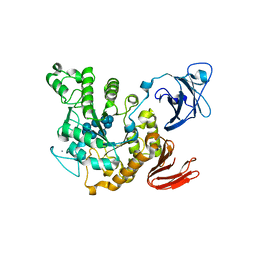 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VTI
 
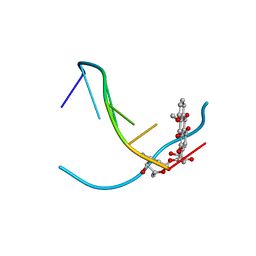 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURES OF D(TGATCA) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Nunn, C.M, Van Meervelt, L, Zhang, S, Moore, M.H, Kennard, O. | | Deposit date: | 1992-03-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DNA-Drug Interactions: The Crystal Structures of d(TGTACA) and d(TGATCA) Complexed with Daunomycin
J.Mol.Biol., 222, 1991
|
|
282D
 
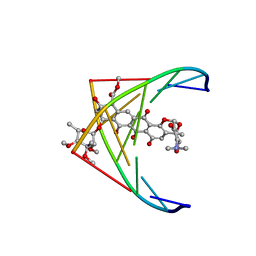 | | A CONTINOUS TRANSITION FROM A-DNA TO B-DNA IN THE 1:1 COMPLEX BETWEEN NOGALAMYCIN AND THE HEXAMER DCCCGGG | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), NOGALAMYCIN | | Authors: | Cruse, W, Saludjian, P, Leroux, Y, Leger, Y, El Manouni, D, Prange, T. | | Deposit date: | 1996-08-26 | | Release date: | 1996-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A continuous transition from A-DNA to B-DNA in the 1:1 complex between nogalamycin and the hexamer dCCCGGG.
J.Biol.Chem., 271, 1996
|
|
2D0H
 
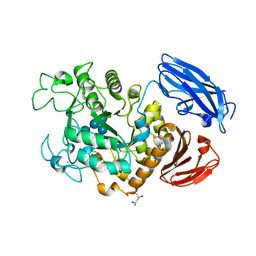 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2VCD
 
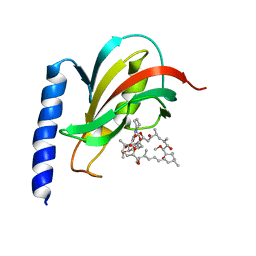 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
1YFC
 
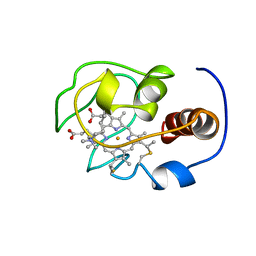 | | Solution nmr structure of a yeast iso-1-ferrocytochrome C | | Descriptor: | HEME C, YEAST ISO-1-FERROCYTOCHROME C | | Authors: | Baistrocchi, P, Banci, L, Bertini, I, Turano, P, Bren, K.L, Gray, H.B. | | Deposit date: | 1996-08-08 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c.
Biochemistry, 35, 1996
|
|
1JR1
 
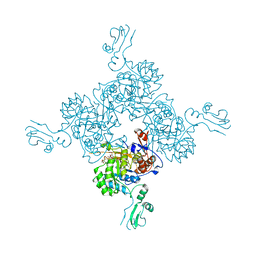 | | Crystal structure of Inosine Monophosphate Dehydrogenase in complex with Mycophenolic Acid | | Descriptor: | INOSINIC ACID, Inosine-5'-Monophosphate Dehydrogenase 2, MYCOPHENOLIC ACID, ... | | Authors: | Sintchak, M.D, Fleming, M.A, Futer, O, Raybuck, S.A, Chambers, S.P, Caron, P.R, Murcko, M.A, Wilson, K.P. | | Deposit date: | 2001-08-09 | | Release date: | 2001-09-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid.
Cell(Cambridge,Mass.), 85, 1996
|
|
1KAN
 
 | |
7DBL
 
 | | Acyl-CoA hydrolase MpaH' mutant S139A in complex with MPA | | Descriptor: | MYCOPHENOLIC ACID, acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
2GFI
 
 | | Crystal structure of the phytase from D. castellii at 2.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Hoh, F. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Debaryomyces castellii CBS 2923 phytase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2D2O
 
 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
3A6O
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/acarbose complex | | Descriptor: | ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2009-09-07 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
5FB7
 
 | |
5ZI9
 
 | | Crystal structure of type-II LOG from Streptomyces coelicolor A3 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, ... | | Authors: | Seo, H, Kim, K.-J. | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical characterization of the type-II LOG protein from Streptomyces coelicolor A3.
Biochem. Biophys. Res. Commun., 499, 2018
|
|
4JAX
 
 | | Crystal structure of dimeric KlHxk1 in crystal form X | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Strater, N, Kettner, K, Otto, A, Lilie, H, Golbik, R.P, Kriegel, T.M. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | In vivo phosphorylation and in vitro autophosphorylation-inactivation of Kluyveromyces lactis hexokinase KlHxk1.
Biochem.Biophys.Res.Commun., 435, 2013
|
|
3U75
 
 | | Structure of E230A-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosylnystose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
4KR8
 
 | |
3U14
 
 | | Structure of D50A-fructofuranosidase from Schwanniomyces occidentalis complexed with inulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
6AZ3
 
 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
6AZ1
 
 | | Cryo-EM structure of the small subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | E-site tRNA, LACK1, MAGNESIUM ION, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Matzov, D, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
7N7G
 
 | | Crystal Structure of FosB from Enterococcus faecium with Fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, MANGANESE (II) ION, ... | | Authors: | Shay, M.R, Simmons, Z, Thompson, M.K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of fosfomycin resistance conferred by FosB from Enterococcus faecium.
Protein Sci., 31, 2022
|
|
7KBV
 
 | |
7T1Z
 
 | | Structure of the Fbw7-Skp1-MycNdegron complex | | Descriptor: | F-box/WD repeat-containing protein 7, Myc proto-oncogene N terminal degron, S-phase kinase-associated protein 1, ... | | Authors: | Wang, B, Rusnac, D.V, Clurman, B.E, Zheng, N. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Sci Adv, 8, 2022
|
|
7T1Y
 
 | | Structure of the Fbw7-Skp1-MycCdegron complex | | Descriptor: | F-box/WD repeat-containing protein 7, Myc proto-oncogene protein C terminal degron, S-phase kinase-associated protein 1, ... | | Authors: | Wang, B, Rusnac, D.V, Clurman, B.E, Zheng, N. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Sci Adv, 8, 2022
|
|
