8AO1
 
 | | solution structure of nanoFAST fluorogen-activating protein in the apo state | | Descriptor: | nanoFAST | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
8AO0
 
 | | Solution structure of nanoFAST/HBR-DOM2 complex | | Descriptor: | (5~{Z})-5-[(2,5-dimethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Photoactive yellow protein | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baleeva, N.S, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
7CVN
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X, Zhou, J, Xu, B. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
8AVV
 
 | | Cryo-EM structure of DrBphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
8AVX
 
 | |
8AVW
 
 | | Cryo-EM structure of DrBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
8ASB
 
 | | Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8ASG
 
 | | Structure of the SFTSV L protein bound in a resting state [RESTING] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*AP*UP*GP*A*)-3'), RNA (5'-R(*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U*)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8ASD
 
 | | Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS7
 
 | | Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS6
 
 | | Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK] | | Descriptor: | RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*C)-3'), RNA-dependent RNA-polymerase L protein | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
6EB6
 
 | | Crystal structure of BAX W139A monomer | | Descriptor: | Apoptosis regulator BAX, FORMIC ACID | | Authors: | Robin, A.Y. | | Deposit date: | 2018-08-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | BAX Activation: Mutations Near Its Proposed Non-canonical BH3 Binding Site Reveal Allosteric Changes Controlling Mitochondrial Association.
Cell Rep, 27, 2019
|
|
4PZN
 
 | | Crystal structure of PHC3 SAM L971E | | Descriptor: | 1,2-ETHANEDIOL, Polyhomeotic-like protein 3 | | Authors: | Nanyes, D.R, Junco, S.E, Taylor, A.B, Robinson, A.K, Patterson, N.L, Shivarajpur, A, Halloran, J, Hale, S.M, Kaur, Y, Hart, P.J, Kim, C.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif.
Proteins, 82, 2014
|
|
7CP2
 
 | | Crystal structure of the African swine fever virus core shell protein p15 | | Descriptor: | CP530R | | Authors: | Liu, K.F, Meng, Y.M, Chai, Y, Li, L.J, Sun, H, Gao, G.F, Tan, S.G, Qi, J.X. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the African swine fever virus core shell protein p15
Biosaf Health, 2021
|
|
5BWZ
 
 | |
7CV0
 
 | | Crystal structure of B. halodurans NiaR in apo form | | Descriptor: | Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
4R4M
 
 | | Crystal structure of C42L cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
6EI6
 
 | | CC2D1B coordinates ESRCT-III activity during the mitotic reformation of the nuclear envelope | | Descriptor: | Coiled-coil and C2 domain-containing protein 1-like, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Ventimiglia, L.N, Cuesta-Geijo, M.A, Martinelli, N, Caballe, A, Macheboeuf, P, Miguet, N, Parnham, I.M, Olmos, Y, Carlton, J.G, Weissehorn, W, martin-Serrano, J. | | Deposit date: | 2017-09-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | CC2D1B Coordinates ESCRT-III Activity during the Mitotic Reformation of the Nuclear Envelope.
Dev. Cell, 47, 2018
|
|
1X76
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-697 | | Descriptor: | 5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-CARBONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
7WVB
 
 | | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1 | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state
To Be Published
|
|
4L50
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-6-methyl-4-oxoheptyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4L87
 
 | |
8AFZ
 
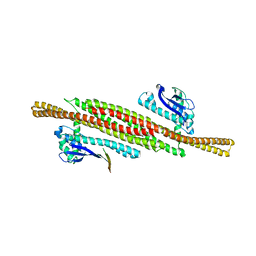 | | Architecture of the ESCPE-1 membrane coat | | Descriptor: | Cation-independent mannose-6-phosphate receptor, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E, Ishida, M, Williamsom, C.D, Banos-Mateos, S, Gil-Carton, D, Romero, M, Vidaurrazaga, A, Fernandez-Recio, J, Rojas, A.L, Bonifacino, J.S, Castano-Diez, D, Hierro, A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4LE1
 
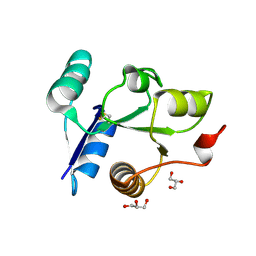 | | Crystal structure of the receiver domain of DesR in the inactive state | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
