4SGB
 
 | |
4CMM
 
 | | Structure of human CD47 in complex with human Signal Regulatory Protein (SIRP) alpha v1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Lea, S.M, Johnson, S, Barclay, A.N. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Polymorphisms in the Human Inhibitory Signal-Regulatory Protein Alpha Do not Affect Binding to its Ligand Cd47.
J.Biol.Chem., 289, 2014
|
|
2SNV
 
 | |
4H5X
 
 | |
1MVT
 
 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MVS
 
 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4H71
 
 | | Human Plk1-PBD in complex with Poloxime ((E)-4-(hydroxyimino)-2-isopropyl-5-methylcyclohexa-2,5-dienone) | | Descriptor: | 2-methyl-5-(1-methylethyl)cyclohexa-2,5-diene-1,4-dione 1-oxime, GLYCEROL, Serine/threonine-protein kinase PLK1 | | Authors: | Yin, Z, Rehse, P.H. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Thymoquinone Blocks pSer/pThr Recognition by Plk1 Polo-Box
Domain As a Phosphate Mimic
Acs Chem.Biol., 8, 2013
|
|
3LOY
 
 | | Crystal structure of a Copper-containing benzylamine oxidase from Hansenula Polymorpha | | Descriptor: | COPPER (II) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Klema, V.J, Johnson, B.J, Wilmot, C.M. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha.
Biochemistry, 49, 2010
|
|
2OQE
 
 | |
2OOV
 
 | |
2QMT
 
 | | Crystal Polymorphism of Protein GB1 Examined by Solid-state NMR and X-ray Diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOPROPYL ALCOHOL, Immunoglobulin G-binding protein G, ... | | Authors: | Frericks Schmidt, H.L, Sperling, L.J, Gao, Y.G, Wylie, B.J, Boettcher, J.M, Wilson, S.R, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Polymorphism of Protein GB1 Examined by Solid-State NMR Spectroscopy and X-ray Diffraction.
J.Phys.Chem.B, 111, 2007
|
|
2P0U
 
 | | crystal structure of Marchantia polymorpha stilbenecarboxylate synthase 2 (STCS2) | | Descriptor: | GLYCEROL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Borel, F, Schroeder, G, Schroeder, J, Ferrer, J.-L. | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | crystal structure of Marchantia polymorpha stilbenecarboxylate synthase 2 (STCS2)
To be Published
|
|
1XP7
 
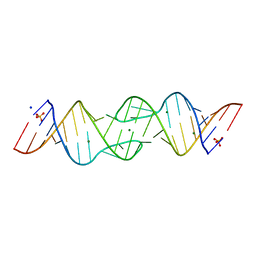 | | HIV-1 subtype F genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XPF
 
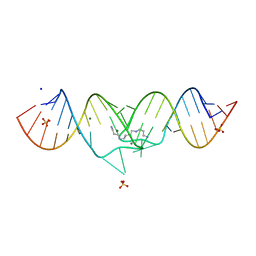 | | HIV-1 subtype A genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XPE
 
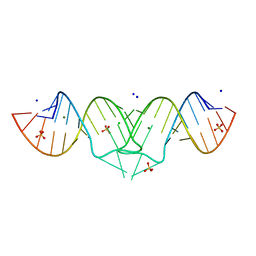 | | HIV-1 subtype B genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
2CI2
 
 | |
1Y3O
 
 | | HIV-1 DIS RNA subtype F- Mn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, SODIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-11-26 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
1Y3S
 
 | | HIV-1 DIS RNA subtype F- MPD form | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-11-26 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
2QXG
 
 | | Crystal Structure of Human Kallikrein 7 in Complex with Ala-Ala-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, L-alanyl-N-[(1S,2R)-1-benzyl-2-hydroxypropyl]-L-alaninamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Bode, W, Steiner, T, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QXH
 
 | | Crystal Structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QXJ
 
 | | Crystal Structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone and Copper | | Descriptor: | COPPER (II) ION, Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QXI
 
 | | High resolution structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RHP
 
 | | The Thrombospondin-1 Polymorphism Asn700Ser Associated with Cornoary Artery Disease Causes Local and Long-Ranging Changes in Protein Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Carlson, C.B, Keck, J.L, Mosher, D.F. | | Deposit date: | 2007-10-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Influences of the N700S Thrombospondin-1 Polymorphism on Protein Structure and Stability.
J.Biol.Chem., 283, 2008
|
|
1ZCI
 
 | | HIV-1 DIS RNA subtype F- monoclinic form | | Descriptor: | 5'-R(*CP*(5BU)P*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', POTASSIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
1YXP
 
 | | HIV-1 DIS RNA subtype F- Zn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', ZINC ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-02-22 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
