5FQ7
 
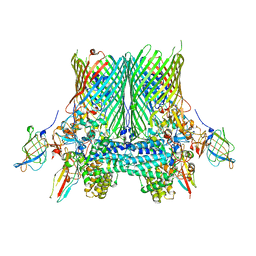 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2261, BT_2262, BT_2263, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FWL
 
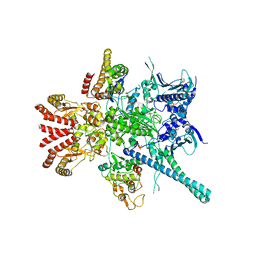 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FQS
 
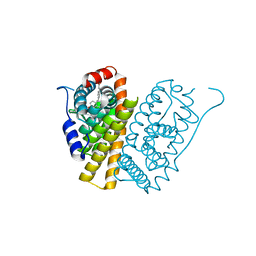 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 3. | | Descriptor: | (E)-3-[4-(6-HYDROXY-2-ISOBUTYL-1-METHYL-3,4-DIHYDROISOQUINOLIN-1-YL)PHENYL]PROP-2-ENOIC ACID, ESTROGEN RECEPTOR | | Authors: | Scott, J.S, Bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
8CJA
 
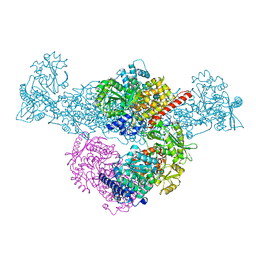 | | A225L/F231A variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5FZE
 
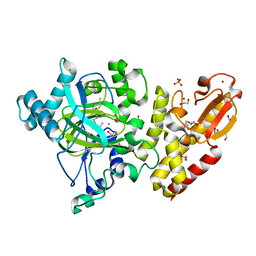 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC3960 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Rotili, D, Mai, A, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc3960
To be Published
|
|
4XTC
 
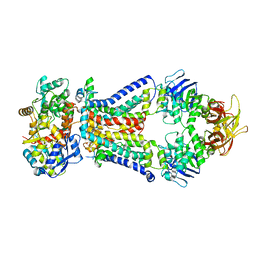 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
1UH1
 
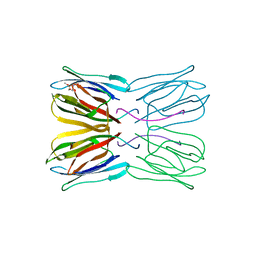 | | Crystal structure of jacalin- GalNAc-beta(1-3)-Gal-alpha-O-Me complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-methyl alpha-D-galactopyranoside, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1TP8
 
 | | CRYSTAL STRUCTURE OF A GALACTOSE SPECIFIC LECTIN FROM ARTOCARPUS HIRSUTA IN COMPLEX WITH METHYL-a-D-GALACTOSE | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA CHAIN, methyl alpha-D-galactopyranoside | | Authors: | Rao, K.N, Suresh, C.G, Katre, U.V, Gaikwad, S.M, Khan, M.I. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two orthorhombic crystal structures of a galactose-specific lectin from Artocarpus hirsuta in complex with methyl-alpha-D-galactose.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7SAB
 
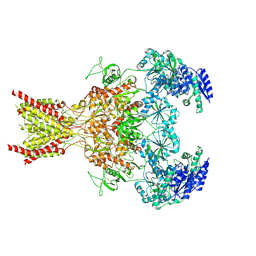 | | Phencyclidine-bound GluN1a-GluN2B NMDA receptors | | Descriptor: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SAD
 
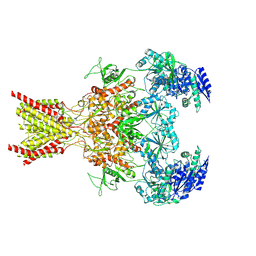 | | Memantine-bound GluN1a-GluN2B NMDA receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SAA
 
 | |
6WJR
 
 | |
1B0O
 
 | |
7CMU
 
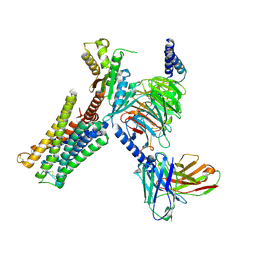 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7CMV
 
 | | Dopamine Receptor D3R-Gi-PD128907 complex | | Descriptor: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
4A7M
 
 | | cytochrome c peroxidase S81W mutant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
4A78
 
 | | cytochrome c peroxidase M119W in complex with guiacol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, Guaiacol, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-11 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
8HHP
 
 | |
6TXZ
 
 | | FAB PART OF M6903 IN COMPLEX WITH HUMAN TIM3 | | Descriptor: | Fab H, Fab L, Hepatitis A virus cellular receptor 2 | | Authors: | Musil, D, Sood, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Identification and characterization of M6903, an antagonistic anti-TIM-3 monoclonal antibody.
Oncoimmunology, 9, 2020
|
|
4A71
 
 | | cytochrome c peroxidase in complex with phenol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PHENOL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
1UCB
 
 | |
6WJS
 
 | |
6GRN
 
 | | CELLOBIOHYDROLASE I (CEL7A) FROM Trichoderma reesei with S-dihydroxypropranolol in the active site | | Descriptor: | 2-[[(2~{S})-3-naphthalen-1-yloxy-2-oxidanyl-propyl]amino]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Sandgren, M, Fagerstrom, A, Widmalm, G, Stahlberg, J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enantioselective Binding of Propranolol and Analogues Thereof to Cellobiohydrolase Cel7A.
Chemistry, 24, 2018
|
|
1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1BV8
 
 | |
