5UF3
 
 | |
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
7TXT
 
 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | Descriptor: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | Authors: | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | Deposit date: | 2022-02-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AM6
 
 | |
8AKW
 
 | | 280 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
5TT3
 
 | |
4YHA
 
 | |
8AAV
 
 | |
7TXD
 
 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BON
 
 | |
4YLI
 
 | | CL-K1 trimer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collectin-11, ... | | Authors: | Wallis, R, Girija, U.V, Gingras, A.R, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis of sugar recognition by collectin-K1 and the effects of mutations associated with 3MC syndrome.
Bmc Biol., 13, 2015
|
|
7H6V
 
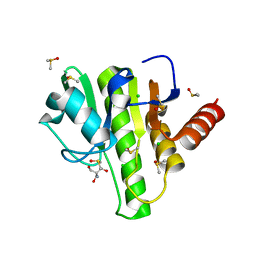 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1262633486 (CHIKV_MacB-x0394) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(difluoromethyl)-1-methyl-1H-pyrazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
5TR6
 
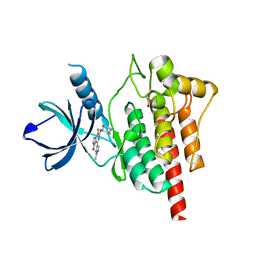 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(1R,2S)-2-aminocyclohexyl]amino}-7-fluoro-4-(1-methyl-1H-pyrazol-4-yl)-1,2-dihydro-3H-pyrrolo[3,4-c]pyridin-3-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
7TPI
 
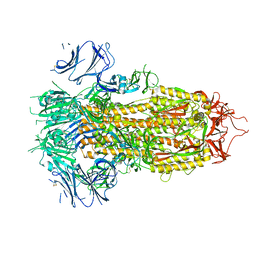 | | SARS-CoV-2 E406W mutant Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Addetia, A, Veesler, D. | | Deposit date: | 2022-01-25 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | SARS-CoV-2 E406W mutant Spike ectodomain
To Be Published
|
|
7H88
 
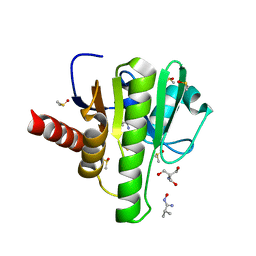 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1741972444 (CHIKV_MacB-x1134) | | Descriptor: | (1Z)-N-hydroxy-2-methylpropanimidamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H8I
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z362772344 (CHIKV_MacB-x1194) | | Descriptor: | 1,4,6-trimethyl-1,2-dihydro-3H-pyrazolo[3,4-b]pyridin-3-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H8W
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z19674820 (CHIKV_MacB-x1316) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7UKM
 
 | |
7UKL
 
 | |
4Z3W
 
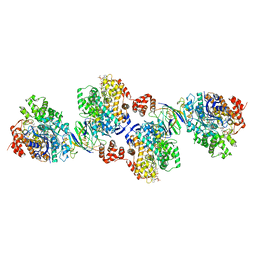 | | Active site complex BamBC of Benzoyl Coenzyme A reductase in complex with 1,5 Dienoyl-CoA | | Descriptor: | 1,5 Dienoyl-CoA, Benzoyl-CoA reductase, putative, ... | | Authors: | Weinert, T, Kung, J, Weidenweber, S, Huwiler, S, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
8BFL
 
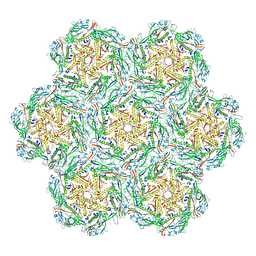 | | Jumbo Phage phi-kp24 empty capsid hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
5U84
 
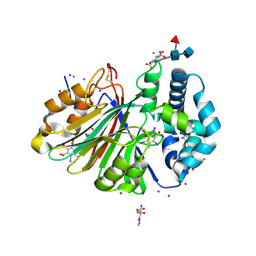 | | Acid ceramidase (ASAH1, aCDase) from common minke whale, Cys143Ala, uncleaved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ... | | Authors: | Gebai, A, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the activation of acid ceramidase.
Nat Commun, 9, 2018
|
|
5U8U
 
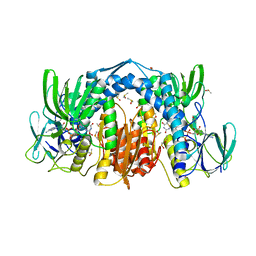 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
5UGF
 
 | | Crystal structure of human purine nucleoside phosphorylase (F159Y) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Cameron, S.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-01-08 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic-site design for inverse heavy-enzyme isotope effects in human purine nucleoside phosphorylase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
