3THJ
 
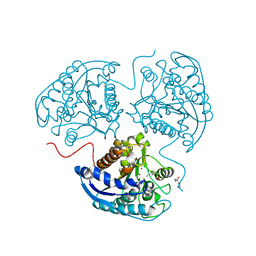 | |
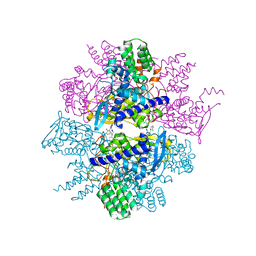
4LFE
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, partially liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl diphosphate synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
3TG6
 
 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
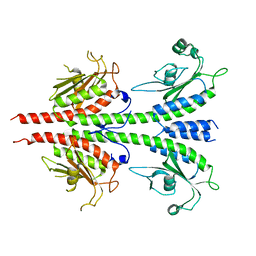
2HZ4
 
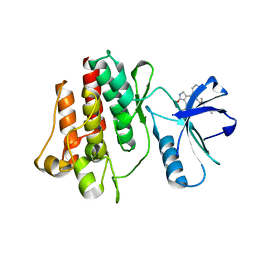 | | Abl kinase domain unligated and in complex with tetrahydrostaurosporine | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3MF0
 
 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
3TPC
 
 | | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021 | | Descriptor: | Short chain alcohol dehydrogenase-related dehydrogenase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021
To be Published
|
|
5TW1
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with RbpA | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Hubin, E.A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA.
Elife, 6, 2017
|
|
3STP
 
 | | Crystal structure of a putative galactonate dehydratase | | Descriptor: | Galactonate dehydratase, putative, MAGNESIUM ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-11 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a putative galactonate dehydratase
To be Published
|
|
2PV7
 
 | |
1PU7
 
 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
4J2U
 
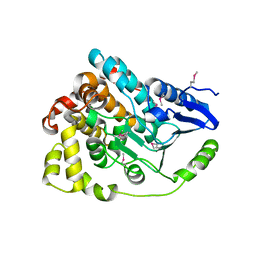 | | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1 | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1
To be Published
|
|
3MBA
 
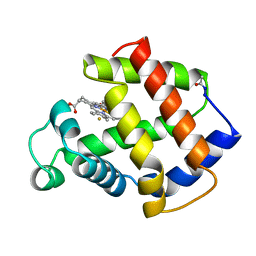 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | FLUORIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
3T8Q
 
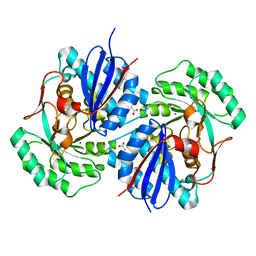 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica | | Descriptor: | MAGNESIUM ION, MALONATE ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica
To be Published
|
|
1V0P
 
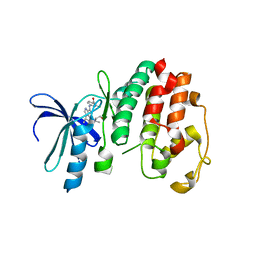 | | Structure of P. falciparum PfPK5-Purvalanol B ligand complex | | Descriptor: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG, PURVALANOL B | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
2A8G
 
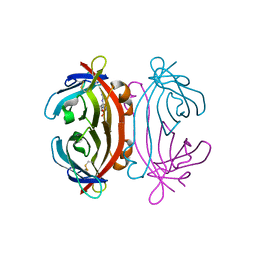 | | Structure of Avidin in complex with the ligand deoxyguanosine | | Descriptor: | 2'-DEOXY-GUANOSINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-07-08 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
4E1J
 
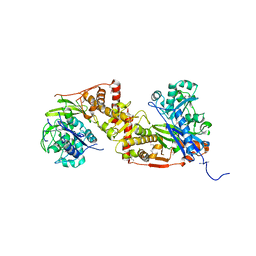 | | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol kinase, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021
To be Published
|
|
4DGS
 
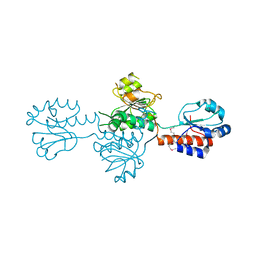 | | The crystals structure of dehydrogenase from Rhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystals structure of dehydrogenase from Rhizobium meliloti
To be Published
|
|
4DYV
 
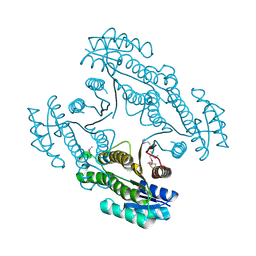 | | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2 | | Descriptor: | CHLORIDE ION, Short-chain dehydrogenase/reductase SDR | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Gizzi, A, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2
To be Published
|
|
2GJH
 
 | |
4IA6
 
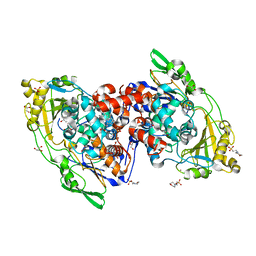 | | Hydratase from lactobacillus acidophilus in a ligand bound form (LA LAH) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HY3
 
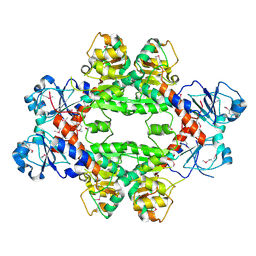 | | Crystal structure of a phosphoglycerate oxidoreductase from rhizobium etli | | Descriptor: | phosphoglycerate oxidoreductase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a phosphoglycerate oxidoreductase from rhizobium etli
To be Published
|
|
4IA7
 
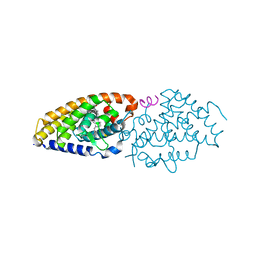 | | Diastereotopic and Deuterium Effects in Gemini | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Maehr, H, Rochel, N, Suh, N, Uskokovic, M. | | Deposit date: | 2012-12-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Diastereotopic and deuterium effects in gemini.
J.Med.Chem., 56, 2013
|
|
2B1N
 
 | |
4IA1
 
 | | Diastereotopic and Deuterium Effects in Gemini | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Maehr, H, Rochel, N, Suh, N, Uskokovic, M. | | Deposit date: | 2012-12-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Diastereotopic and deuterium effects in gemini.
J.Med.Chem., 56, 2013
|
|
4DU5
 
 | | Crystal structure of PfkB protein from Polaromonas sp. JS666 | | Descriptor: | CHLORIDE ION, PfkB | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hellerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of PfkB protein from Polaromonas sp. JS666
To be Published
|
|
